Kallol Gupta
@GuptaLab1
We make tools that render a quantitative view of #membraneproteins and #lipids at the #cellmembrane @YaleCellBio. #Massspectrometers are our favorite toys
你可能會喜歡
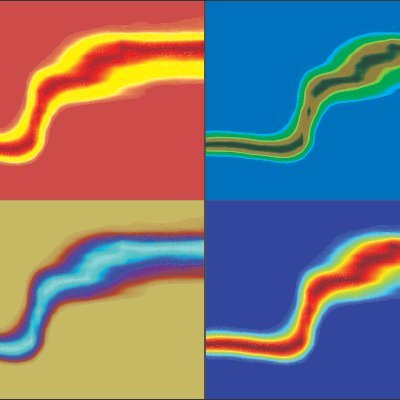
Excited to see our work on the cover of @naturemethods We present a #nativemassspectrometry platform to detect #membraneprotein #lipid complexes directly from tunable #membranes Special thanks to Hannah Wang. One for #TeamMassSpec @YaleCellBio @YaleWestCampus @YaleMed @Yale
Our June issue is live! 🎉 nature.com/nmeth/volumes/… On the cover, from @GuptaLab1 and artist Hanna Wang, a native mass spectrometry platform captures membrane protein–lipid organization directly from tunable lipid membranes. Paper here: nature.com/articles/s4159…

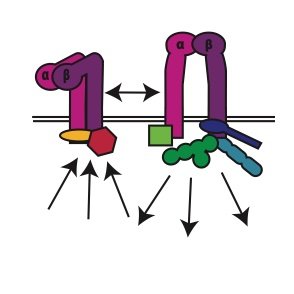
If @ #BPS2025 , check out the talk by @CBcellbiologist today in the "Protein Lipid Interactions: Structure and Dynamics" Session @ 12PM. She will present our recently developed platform to capture membrane proteins in their native nanodomains & some exciting applications of it
If you are @ProteinSociety annual meeting, check out the talks and posters by @CBcellbiologist and @Rachelmcalli on our new tools to understand how the native #membrane regulates #membraneproteins and its application on a B cell protein (with @moitrayee_lab)

If you are @ #BPS #Membrane #Biophysics meeting @Tahoe, check out Caroline's talk today. She describes our new & exciting platform to capture >2000 unique #membraneproteins in their native nanodomains with high efficiency and nanoscale spatial resolution

If @ #ASMS2024, check out all our exciting works describing new technologies to study molecular organization of #membraneproteins and #lipids at the #membrane

Meet us @moitrayee_lab at #BPS2024 with posters from @GTWalker7 and @Rachelmcalli and my talk at the #FutureofBiophysics Burroughs Wellcome Fund symposium. We will highlight our work on determining #membraneprotein #organization and #structure directly from native #membranes.

If @ #BPS2024, check out all our exciting works describing new technologies to study molecular organization of #membraneproteins and #lipids directly from the #membrane and its applications in understanding various membrane-associated signaling events

very happy to play our part in the fantastic work led by @moitrayee_lab, in collaboration with the @MuzumdarLab. Work spearheaded by two of our champion grad students @GTWalker7 and @CBcellbiologist
Here is a tweetorial summarizing findings from our recent paper from @YalePharm @YaleMed in @NatureNano by @GTWalker7 and @CBcellbiologist. Dream team @shalgene @Xiangyu_GigiG @MuzumdarLab @GuptaLab1! Also see the N&Vs at nature.com/articles/s4156… by @KrishnanYamuna!
A very exciting application of #ESIBD
Out today in @ScienceMagazine: Seeing glycans attached to proteins and lipids - at the *single molecule level*! 🎉🎉🎉 doi.org/10.1126/scienc… #ScienceResearch #glycotime #teammassspec My 1st #tweetorial🧵(1/n) 👇👇👇

What a cool tool!!
Our article on in vivo force sensors appeared in Science Advances. Thank Yuan Ren, Julien Berro @BerroLab, and Jie Yang! Force redistribution in clathrin-mediated endocytosis revealed by coiled-coil force sensors | Science Advances science.org/doi/full/10.11…

Extremely honored to be recognized as an Emerging Investigator by the American Society for Mass Spectrometry @asmsnews. Credit goes to my amazing lab members for their hard work. Special thanks to my past mentors & present colleagues at @YaleWestCampus @YaleCellBio @YaleMed
Our article on in vivo force sensors appeared in Science Advances. Thank Yuan Ren, Julien Berro @BerroLab, and Jie Yang! Force redistribution in clathrin-mediated endocytosis revealed by coiled-coil force sensors | Science Advances science.org/doi/full/10.11…

Turns out Gas phase is not that bad! Very exciting
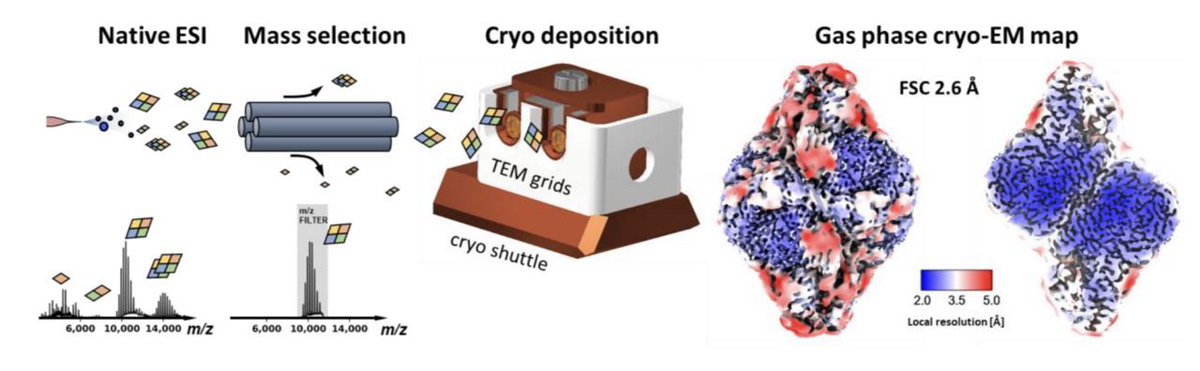
We used #nativeMS electrospray ion beam deposition (ESIBD) to make samples for #cryoEM: mass-filtered, soft-landing deposition, cold substrate, in-situ ice growth, clean+cold transfer...we find... @ESIBDLab with many colleagues @OxfordChemistry @KavliOxford #structuralbiology
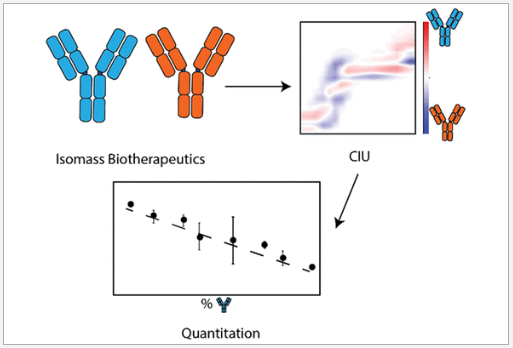
See our lab's latest JASMS publication! In this work, our lab deploys this quantitative CIU methodology to measure isomass pairs of biotherapeutics. Find it here: pubs.acs.org/articlesonrequ… #ASMS #JASMS #CIU #MassSpec #anitbodies
Congrats Justin! Really nice work showing ATG2 function on monolayer-wrapped LDs and careful characterization of a transport dead mutant which loses bridge-like but not shuttle-like lipid transport capacity. Thanks also to @Rachid_Thiam for cool studies with monolayer mimics.
I’m excited to finally share this project from the @TJ_MeliaLab where we demonstrate that the lipid transport protein ATG2 is active and highly efficient on phospholipid monolayers. (1/3) biorxiv.org/content/10.110…
If you are @ProteinSociety meeting #PS37, please check out the exciting work of @anipan84 (Poster20AB), showing how we can directly determine macromolecular organization of #membraneproteins and #lipds directly from synthetic and cellular membranes using #nativemassspectrometry
If you are @ProteinSociety meeting #PS37, please check out the exciting work of our grad student @Rachelmcalli (Poster114AB), showing how lipid vesicle #nativemassspectrometry can decipher membrane recruitment mechanism of a B-cell kinase
Rachel McAlister @Rachelmcalli is representing us at the @ProteinSociety meeting #PS37. Stop by her poster 114AB to check out a native MS based general experimental platform to identify and quantify lipid binding in membrane association domains (collaboration with @GuptaLab1)
Check out our new study published at @NatureComms in which we investigated the Akt1 #signaling and dynamics using an optogenetic-phosphoproteomic system, or Optop-DIA! nature.com/articles/s4146… @YalePharm @CancerYale@YaleWestCampus@yalemed 1/4
United States 趨勢
- 1. Browns 57.6K posts
- 2. Ravens 40.3K posts
- 3. Notre Dame 135K posts
- 4. Bengals 39.4K posts
- 5. Shedeur 59.2K posts
- 6. Stefanski 16K posts
- 7. Josh Allen 18.1K posts
- 8. Steelers 47.6K posts
- 9. Lamar 27.9K posts
- 10. Bills 87.9K posts
- 11. Titans 28.1K posts
- 12. Joe Burrow 13.8K posts
- 13. Colts 29.2K posts
- 14. Saints 32.9K posts
- 15. Bucs 11.6K posts
- 16. #HereWeGo 5,657 posts
- 17. Jordan Love 5,367 posts
- 18. Tulane 40.4K posts
- 19. Tee Higgins 6,168 posts
- 20. Tyler Shough 4,033 posts
Something went wrong.
Something went wrong.