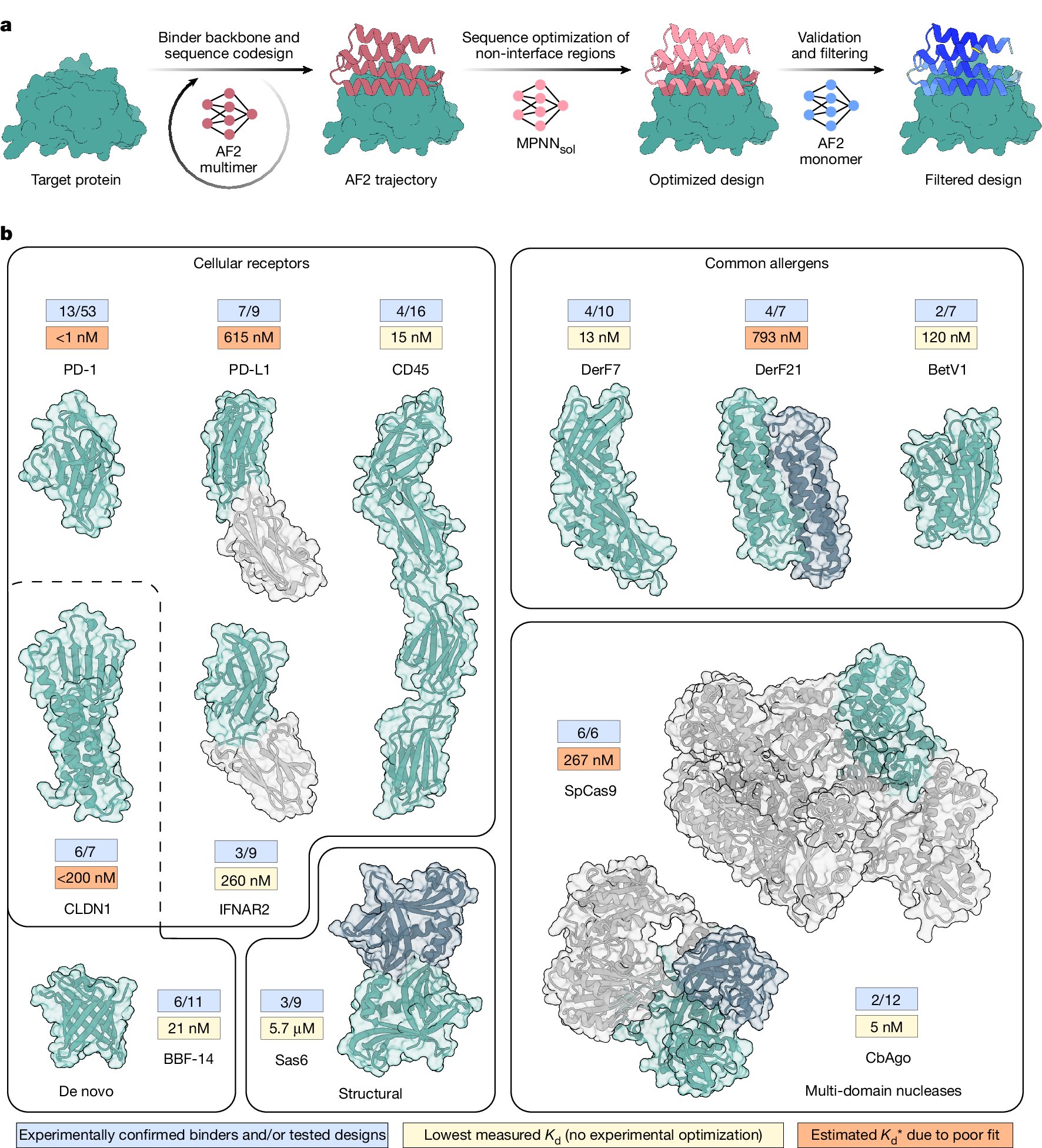
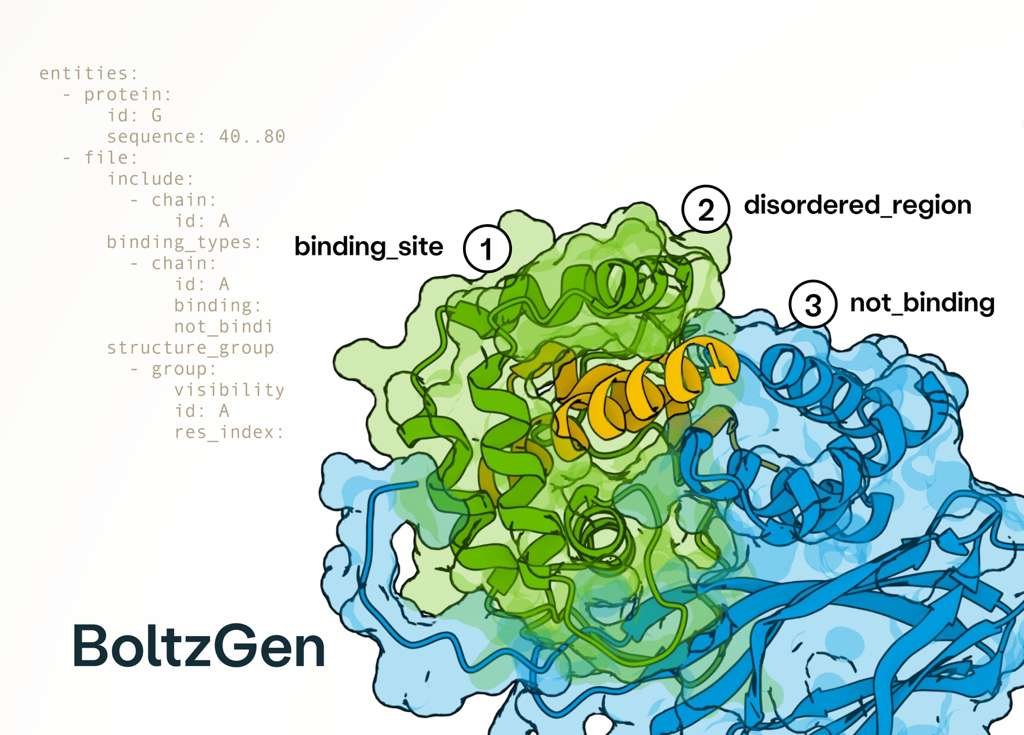
Excited to release BoltzGen which brings SOTA folding performance to binder design! The best part of this project has been collaborating with many leading biologists who tested BoltzGen at an unprecedented scale, showing success on many novel targets and pushing its limits! 🧵..
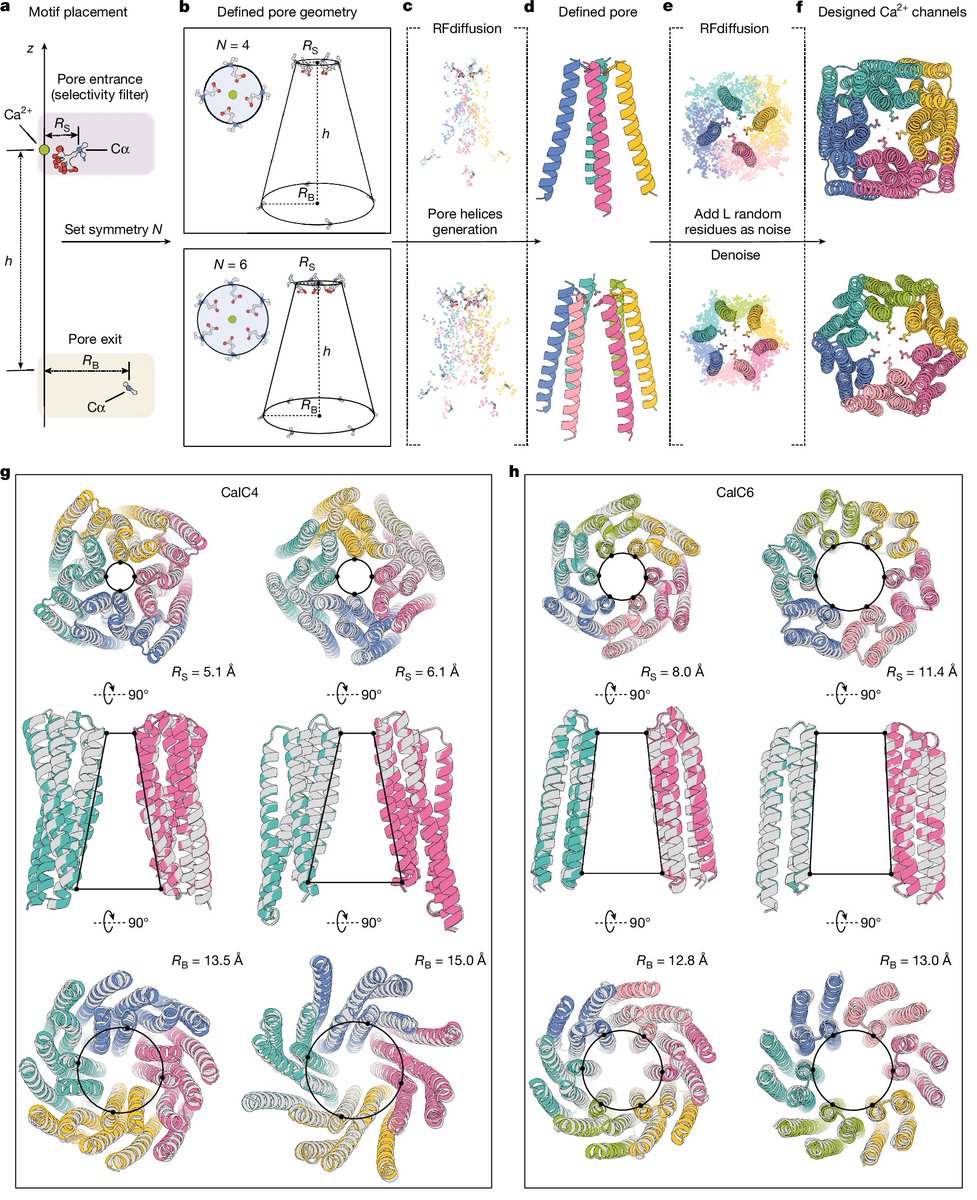
🚀🚀David Baker paper from @Nature is here👇👇 What if we could design calcium-selective ion channels from scratch by precisely positioning atoms at their selectivity filters?@UWproteindesign "Bottom-up design of Ca2+ channels from defined selectivity filter geometry" •…
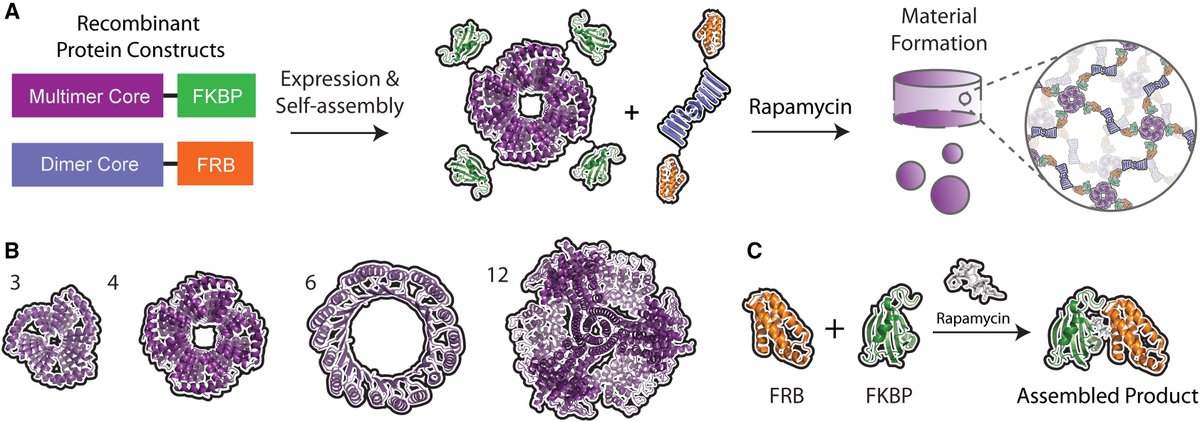
🚀🚀David Baker paper from @CellBiomat is here👇 What if we could design entirely new proteins that self-assemble into customizable materials on command?@UW "Stimuli-triggered formation of de novo-designed protein biomaterials" • Protein-based biomaterials offer advantages…
北川進先生のお言葉。サイエンスの世界は厳しい…。そりゃあ言葉遊びに興じている人文系は負けるわ…。

BREAKING NEWS The 2025 #NobelPrize in Physiology or Medicine has been awarded to Mary E. Brunkow, Fred Ramsdell and Shimon Sakaguchi “for their discoveries concerning peripheral immune tolerance.”

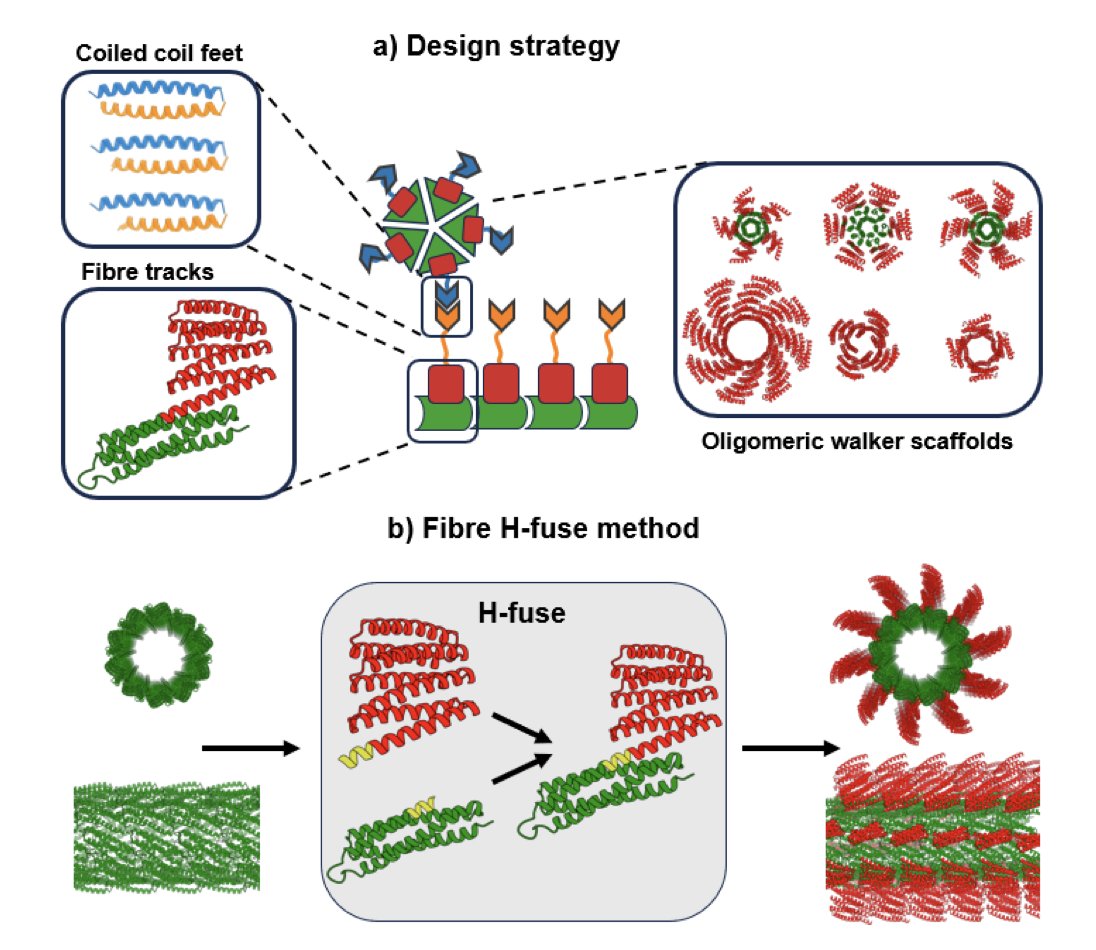
De-novo design of a random protein walker 🚀 New preprint from David Baker!🚀 1. A team of researchers has achieved a significant milestone in protein engineering by designing a random protein walker that diffuses along a designed protein track. This is a major step forward in…
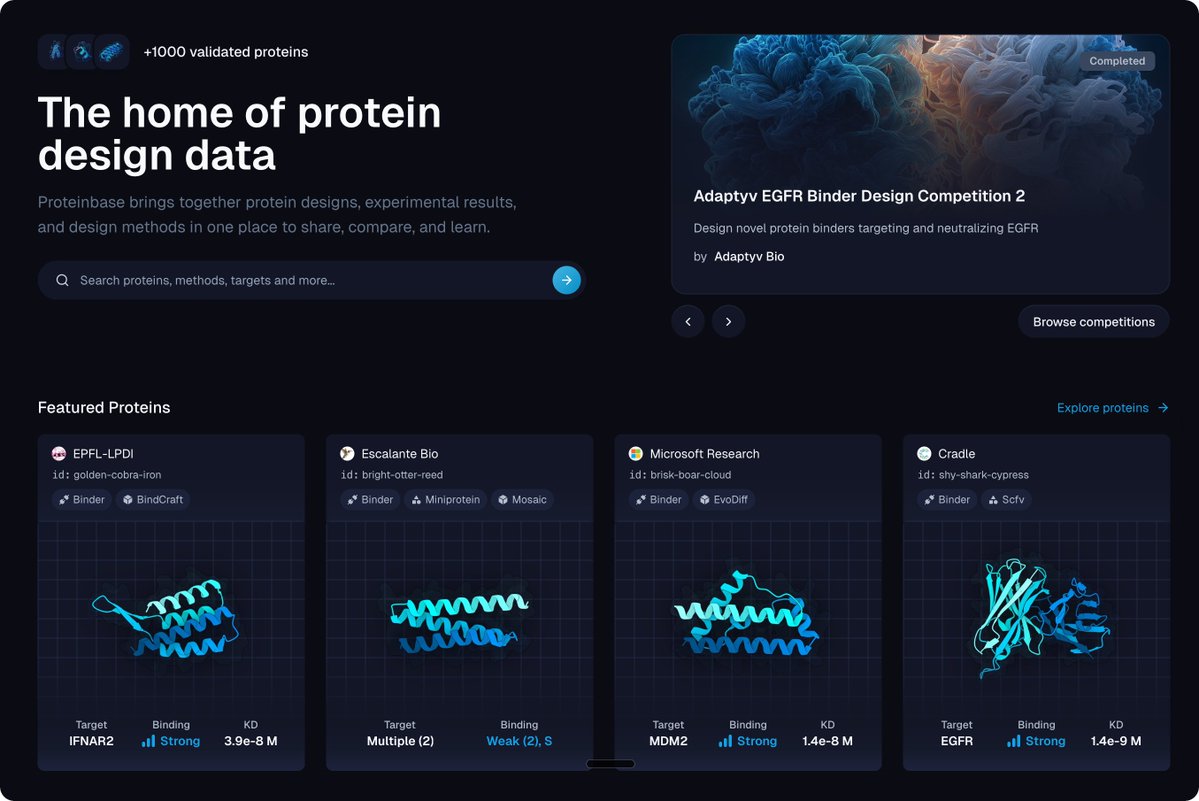
Today we’re launching Proteinbase, a single hub for experimental protein design data. Over 1,000 novel proteins are already live, each with computational predictions, experimental validation, and the method used to design them. Everything comes from one lab under standardized…
🚀🚀David Baker paper is here from @Nature How can scientists engineer proteins to rapidly turn off cellular signaling, like having a molecular "kill switch" for cytokines? @UWproteindesign "Design of facilitated dissociation enables timing of cytokine signalling" • Protein…
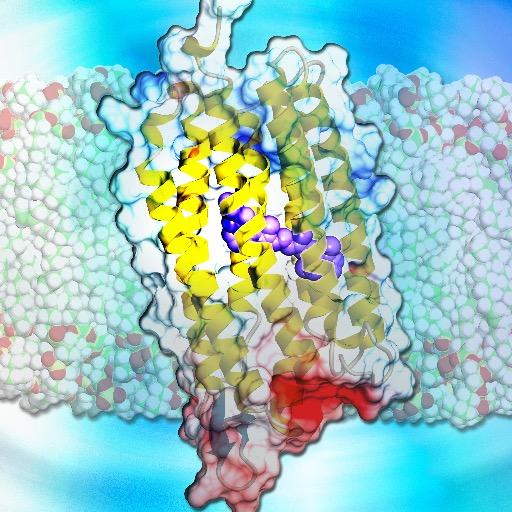
Online now! Substrate and inhibitor binding of human GABA transporter 3 dlvr.it/TMz5hJ
A joint sequence-structure diffusion model for transmembrane proteins!
実験医学増刊号に寄稿させていただきました!深層学習ベースのde novoタンパク質設計でできること&課題を解説しました。 深層学習でゼロからつくる機能性タンパク質【本田信吾】 編集くださった加藤先生 @emeKato 西増先生 @hnisimasu 、編集部の皆様に感謝申し上げます! yodosha.co.jp/jikkenigaku/bo…
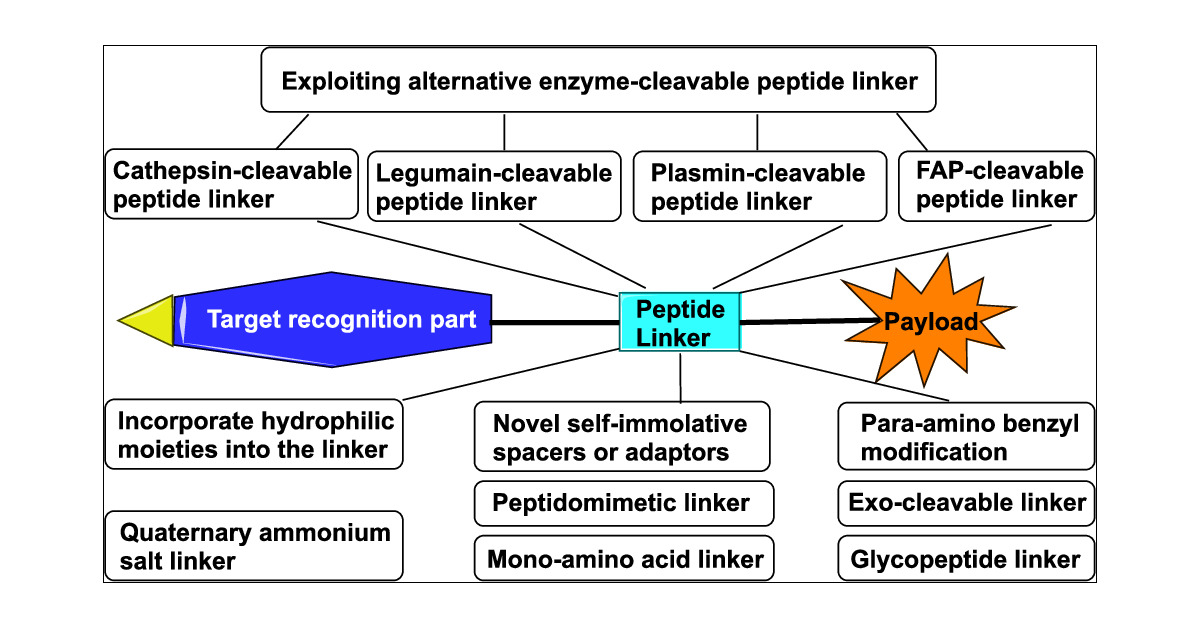
ADCに不可欠なペプチドリンカー(Cleavable linker)についてまとめた総説 pubs.acs.org/doi/10.1021/ac… 非常によくまとまっており、Cleavable linkerを俯瞰されたい方はとりあえずこれを読んでおけばOKかと思います。 ADCリンカーの役割…
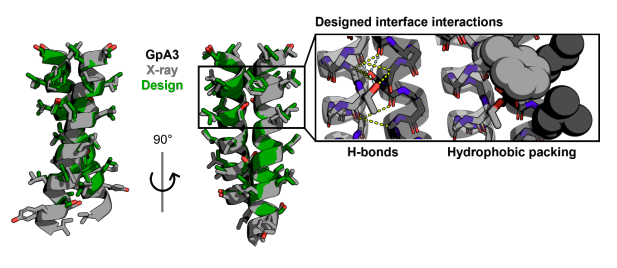
Nature research paper: One-shot design of functional protein binders with BindCraft go.nature.com/4g0R09u
トークの機会をいただきました! 同僚とSummer studentsの活躍によってめちゃアツい展開を迎えているプロジェクトの話をさせていただく予定です!
【超越分子システムセミナーのご案内】10/30(木)の12時からオンラインセミナーを開催します。奮ってご参加いただけますと幸いです! 開催日時:2025年10月30日(木)12:00~13:00 講演者:本⽥ 信吾 先生(ワシントン大学)@akg_entrance、坪山 幸太郎 先生(東京大学)@KotaroTsuboyama

Boltz-2が遅いので、docking結果を突っ込んでaffinity予測させれば速くてそこそこ良い予測ができるよ、というpreprintです。(@yumizsui) [2508.17555] Boltzina: Efficient and Accurate Virtual Screening via Docking-Guided Binding Prediction with Boltz-2 arxiv.org/abs/2508.17555
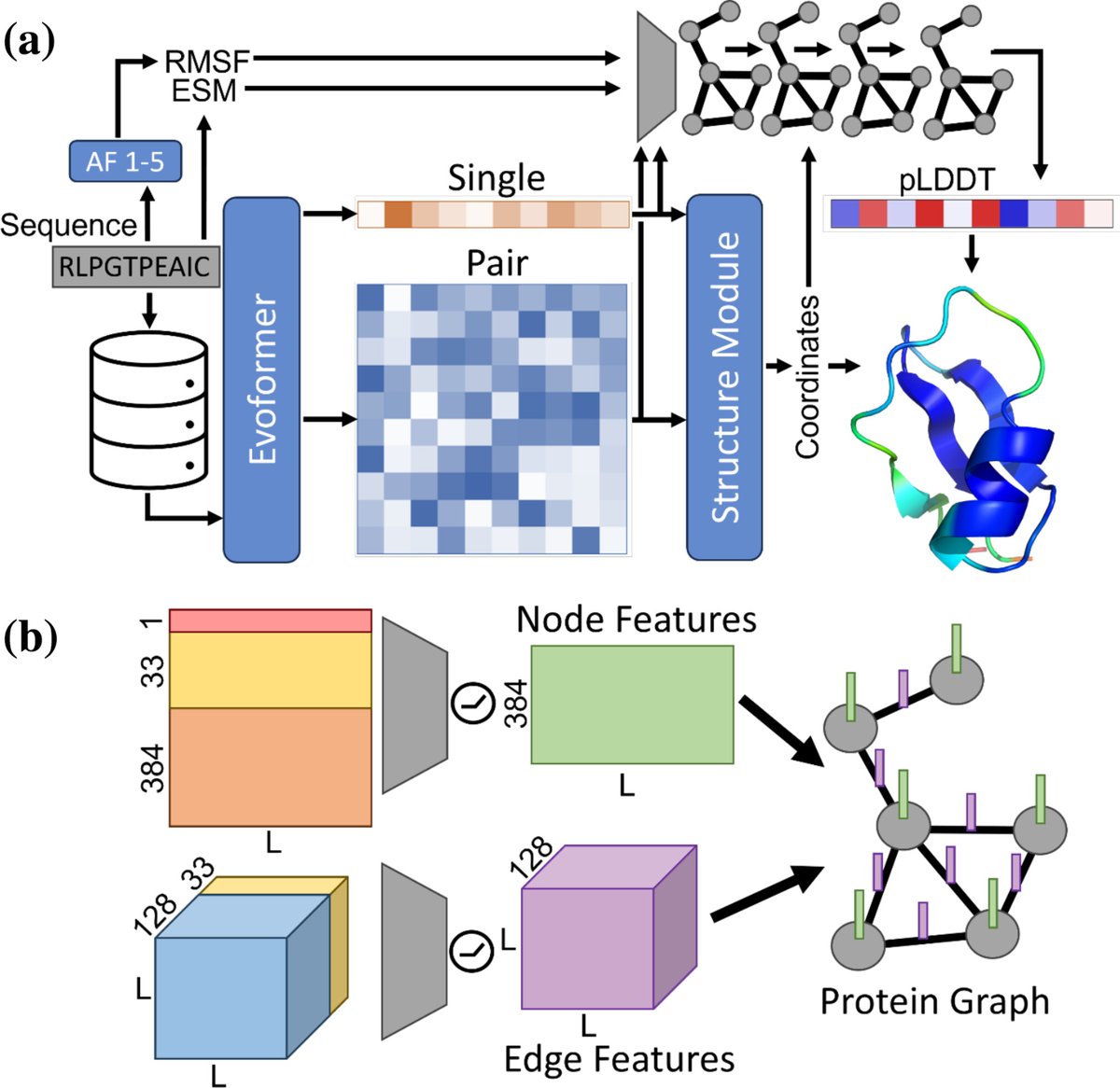
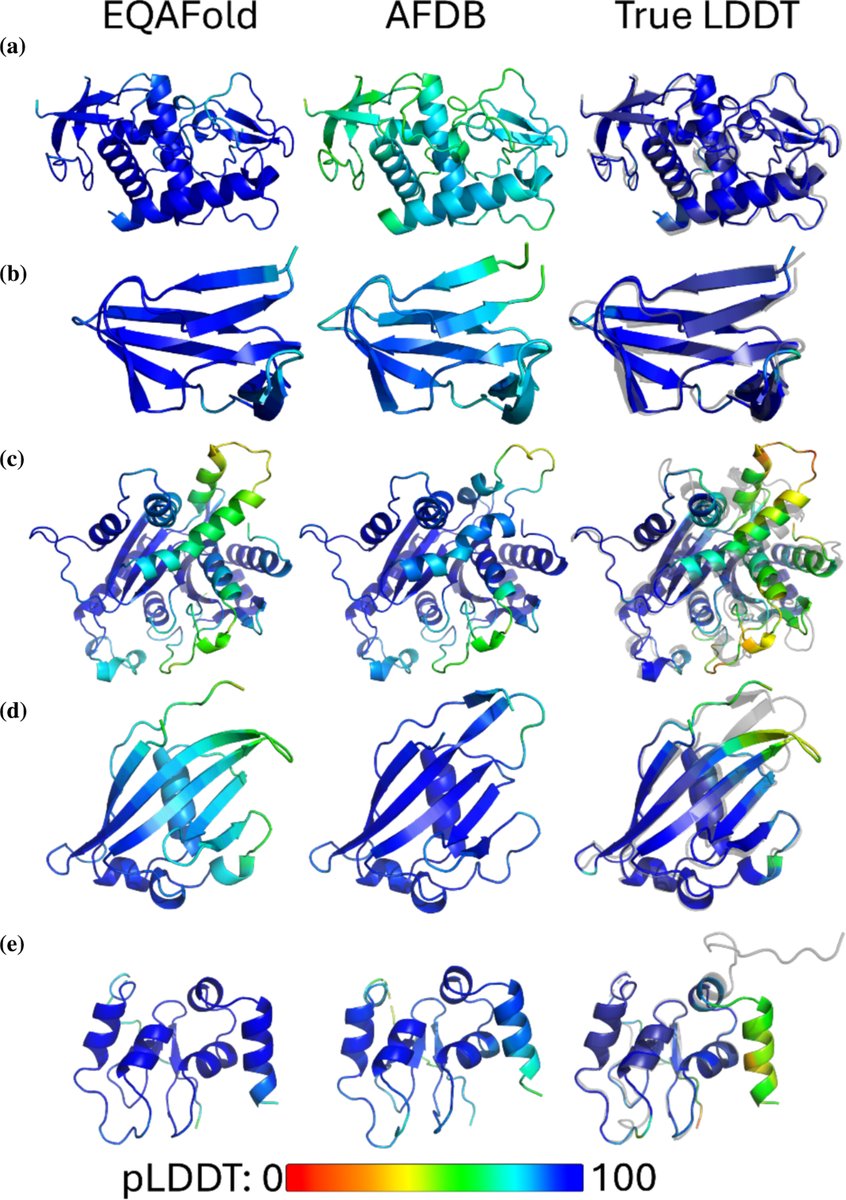
新しい論文出ました。Protein Science. EQAFold! Alphafold2の信頼度スコア(plDDT)は残基単位で見ると実際の正解度と違う、特にスコアが高すぎるケースがあります。ここではAF2にgraph NNを入れることでこれを改善します。
New paper from our lab! Introducing EQAFold: "AlphaFold model quality self‐assessment improvement via deep graph learning" Jacob Verburgt, Zicong Zhang & D. Kihara, Protein Science. onlinelibrary.wiley.com/doi/10.1002/pr…
実験医学の増刊号、「構造生命科学 AlphaFold時代にどう活かす?」に一章書かせていただきました: AlphaFoldを活用したタンパク質複合体構造予測 増刊号の編集していただいた加藤英明,西増弘志の両先生ありがとうございます。 yodosha.co.jp/jikkenigaku/bo…
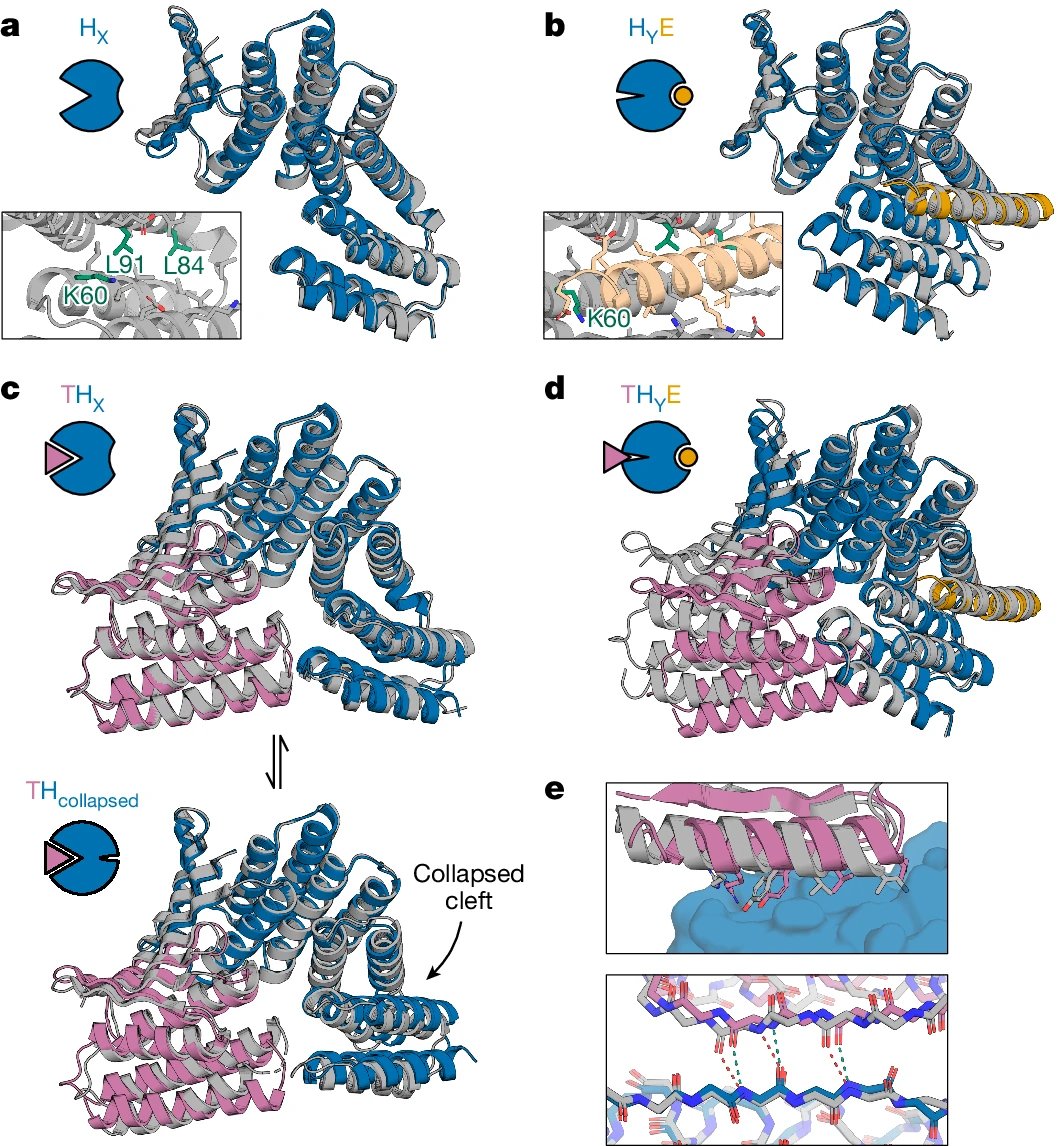
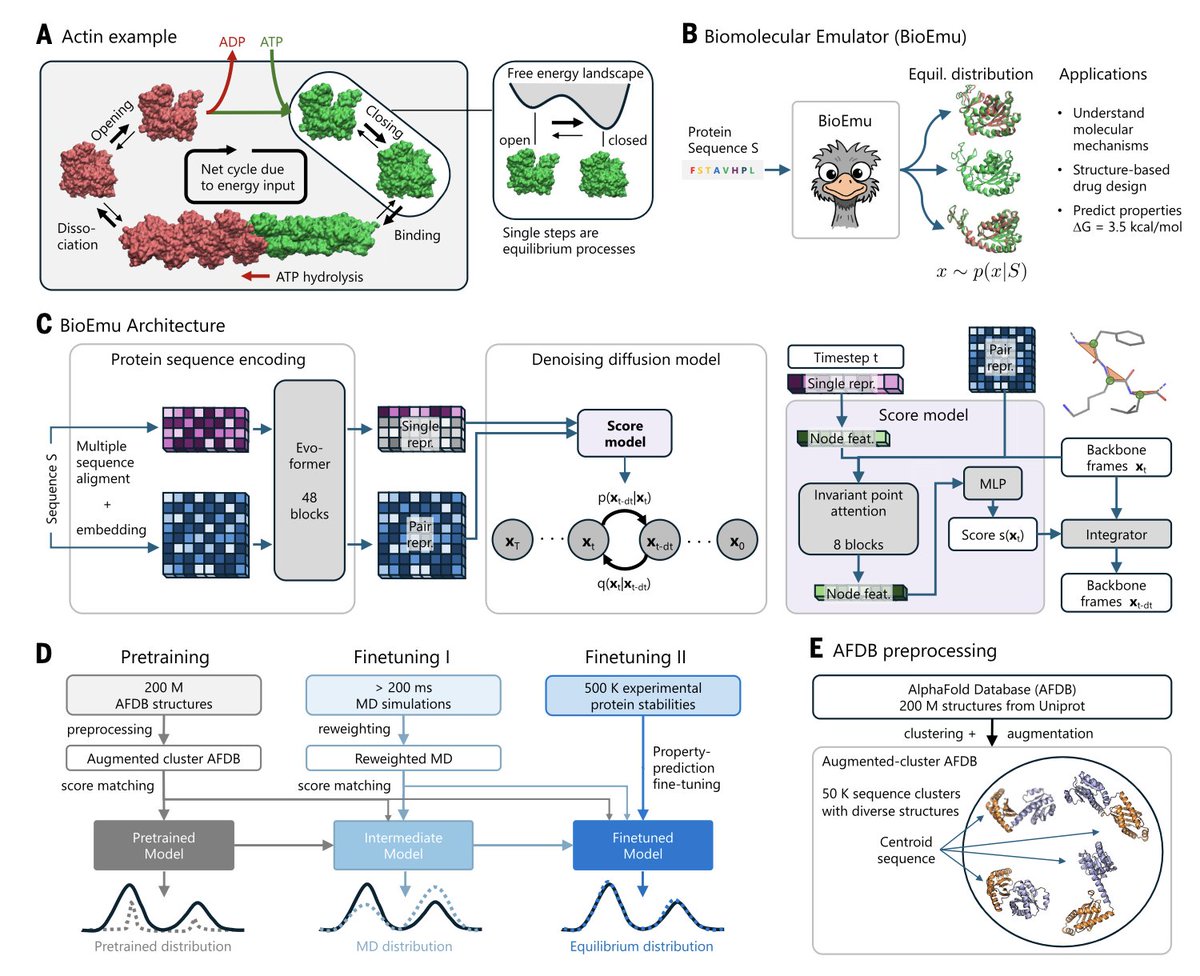
Scalable Emulation of Protein Equilibrium Ensembles with Generative Deep Learning @ScienceMagazine 1. A groundbreaking study introduces BioEmu, a deep learning system that can rapidly generate protein structure ensembles, capturing dynamic conformational changes and predicting…
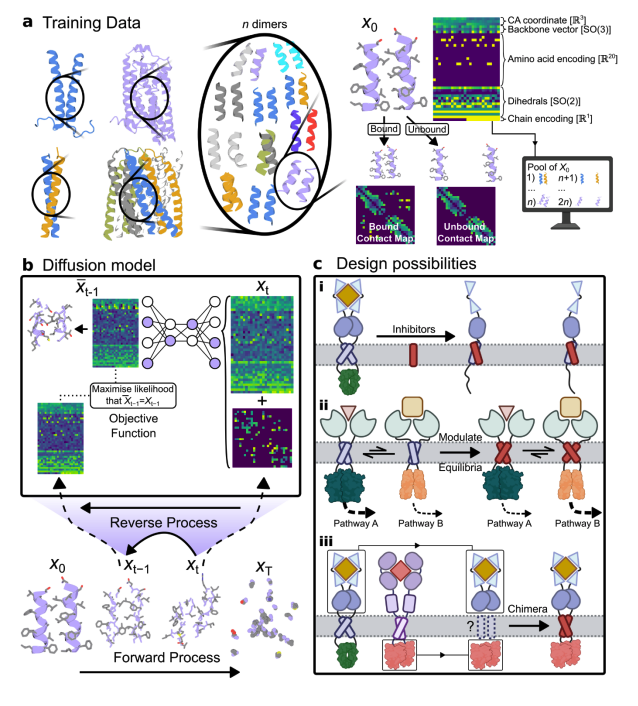
Accelerating Biomolecular Modeling with AtomWorks and RF3 🚀 New preprint from David Baker!🚀 1. A new framework called AtomWorks has been introduced to revolutionize biomolecular modeling. AtomWorks provides a unified and modular platform for developing state-of-the-art…

United States 트렌드
- 1. Austin Reaves 40.6K posts
- 2. Steelers 83.9K posts
- 3. Packers 65.4K posts
- 4. Tomlin 12.2K posts
- 5. Tucker Kraft 15.2K posts
- 6. Jordan Love 16.2K posts
- 7. #GoPackGo 10.6K posts
- 8. Derry 18.9K posts
- 9. #LakeShow 3,630 posts
- 10. #BaddiesAfricaReunion 8,637 posts
- 11. Pretty P 3,701 posts
- 12. Aaron Rodgers 19.6K posts
- 13. #LaGranjaVIP 67.6K posts
- 14. Teryl Austin 2,138 posts
- 15. Dolly 12.1K posts
- 16. Zayne 18.8K posts
- 17. #HereWeGo 7,507 posts
- 18. Sabonis 2,399 posts
- 19. Karola 4,101 posts
- 20. Green Bay 10.9K posts
Something went wrong.
Something went wrong.