SeqCode
@seq_code
Registry: http://seqco.de | SeqCode: http://seqco.de/seqcode | Initiative: http://seqco.de/initiative | Join the SeqCode Community: http://seqco.de/join Public discussion: http://seqco.de/connect
You might like
Alkaliphilic aceticlastic #methanogen isolated from mud volcano. #seqcode #taxonomy #methane frontiersin.org/articles/10.33…
frontiersin.org
Frontiers | Phenotypic and genomic characterization of the first alkaliphilic aceticlastic methan...
Methanogenic enrichments with acetate from a soda lake and a terrestrial mud volcano allowed for the first time to obtain highly purified cultures of alkalip...
Prof. Whitman will be giving an update on the @seq_code at our next #BISMiSLive session this Saturday. Make sure you register at shorturl.at/cqrG6 @Leibniz_DSMZ_en @BccmCollections @CCUG_Sweden @CABI_Microbe @ICSP_news @ari_pune #TaxonomyIsFun

📢 Request for public discussion and ballot to amend #SeqCode rules on the priority of #Candidatus names and correction of typographic and orthographic errors is now available: nature.com/articles/s4370… Stay tuned to join the conversation! @seq_code @ISME_microbes #nomenclature
Final version of the GTDB-defined higher taxa ms is live. doi.org/10.1093/femsle… @ace_gtdb @seq_code #ICNP #nomenclature
If its our inability to grow the so-far uncultivated in the lab, do they not exist? Why differentiate between the ones we can and cannot grow in the lab? #TaxonomyIsFun listen to the debates @BISMiS_ Live YouTube channel @ICSP_news @seq_code
Meet our elephant in the room! We can't hide anymore...so we talk about it a lot :) In this @ASMicrobiology blog, Barny and Brian are discussing why it is essential to link names to genome sequences and what #SeqCode is in short. asm.org/Articles/2023/…
Meet our elephant in the room! We can't hide anymore...so we talk about it a lot :) In this @ASMicrobiology blog, Barny and Brian are discussing why it is essential to link names to genome sequences and what #SeqCode is in short. asm.org/Articles/2023/…
Can we accelerate taxonomy? Check this paper from @sigwartae "...In many cases, it is clear that not all of these data are necessary to distinguish one species from another. This is interesting science, but not actually required for naming species..."
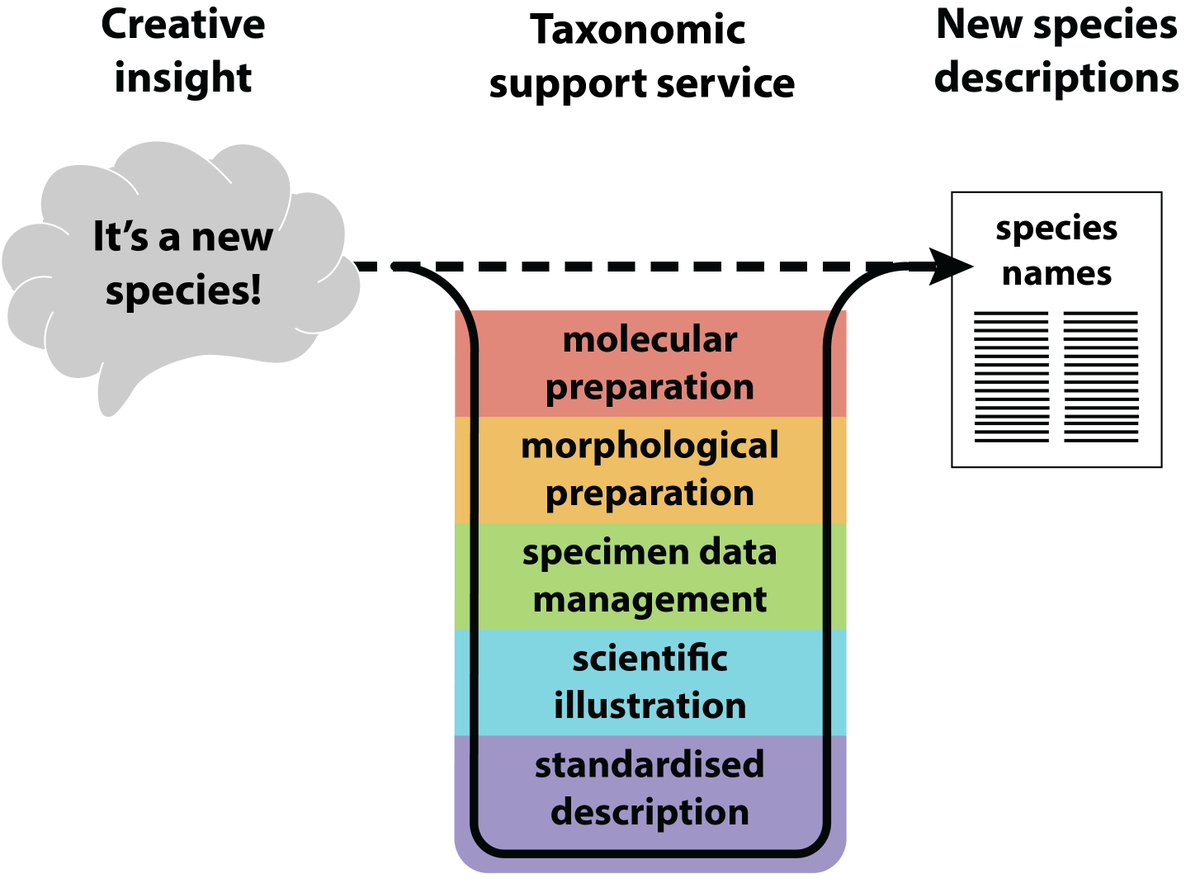
Species are going extinct before we discover them Have you ever wondered, why is there no service to support taxonomic species descriptions? The @oceanspecies_ project @Senckenberg proposes a new model to accelerate taxonomy. Out now in #Bioessays onlinelibrary.wiley.com/doi/10.1002/bi…
Miguel gave a great overview of the #SeqCode and its Registry at #FEMS2023 yesterday. Still got some questions? Don't hesitate to ask or drop your suggestions here: seqcode-public.slack.com/ssb/redirect #seqcode #nomenclature @ISME_microbes
Are you planning to name a new #virus within your bacterial host or both? Consider this new guidelines 👇 "The guidelines presented here focus on the MIUViG standards on genome quality and expand on the naming of sequences and their submission to public databases."
📢Save the date!
Looking forward to this opportunity to share our thoughts @SteenkampEmma @marike_palmer @Fabiteam1 @UPnasagric
Melandre van Lill ready to use the #seqcode to describe her Mesorhizobium species @Fabiteam1 @UPnasagric @seq_code

😍
Promoting the #seqcode for the description of Mesorhizobium species @fanus_venter @SteenkampEmma @Fabiteam1 @UPnasagric @seq_code

📢Dear #community, the #SeqCode Reconciliation Commission is seeking your contribution: please share your opinion on the validation of the species name #Omnitrophus_fodinae by commenting on this request: github.com/seq-code/seqco… Deadline: 28 September 2023

Merci @GilbertGreub for your support! "Members of the ICSP subcommittee for the taxonomy of the phylum Chlamydiae (Chlamydiota)... recommend the registration of new names of uncultured prokaryotes in the SeqCode registry." doi.org/10.1016/j.nmni… #SeqCode @ISME_microbes
Minutes of the last EB meeting has been added to the #ISME website: isme-microbes.org/seqcode-reposi… #SeqCode @ISME_microbes
If you are planning to validate names under different codes, this talk can be useful: How to interpret the #ICNP. Remember that #SeqCode recognise names that are validly published under the ICNP!
#ICNP #TaxonomyIsFun Ever wondered how to interpret and apply the code? Learn all about it from Markus Göker at the next #BISMiSLive session. Register at shorturl.at/dER06 @Leibniz_DSMZ_en @BccmCollections @CCUG_Sweden @ICSP_news @NCMR_Pune @ari_pune @NCIM_NCLPune

A tale of two codes: The continuation ;) researchsquare.com/article/rs-303… #SeqCode #GTDB #ICNP
United States Trends
- 1. Rosalina 19.9K posts
- 2. Bowser Jr 7,052 posts
- 3. Crypto ETFs 2,872 posts
- 4. Jameis 4,373 posts
- 5. #wednesdaymotivation 4,564 posts
- 6. #SuperMarioGalaxyMovie 1,584 posts
- 7. Benny Safdie 3,119 posts
- 8. Michael Wolff 2,067 posts
- 9. H-1B 60.1K posts
- 10. Good Wednesday 32.4K posts
- 11. Hump Day 15.6K posts
- 12. $SENS $0.70 Senseonics CGM N/A
- 13. $LMT $450.50 Lockheed F-35 N/A
- 14. #Talus_Labs N/A
- 15. #Wednesdayvibe 2,431 posts
- 16. Prowler 2,088 posts
- 17. #لماذا_لا_تقبل_الهدنه 3,073 posts
- 18. Captain Marvel 1,710 posts
- 19. $APDN $0.20 Applied DNA N/A
- 20. Operation Dirtbag 1,915 posts
Something went wrong.
Something went wrong.