Dessimoz Lab
@DessimozLab
At the interface between biology and computer science, we seek to better understand evolutionary and functional relationships between genes, genomes & species.
You might like
FastOMA is out now in Nature Methods 🎉: nature.com/articles/s4159… A new orthology inference algorithm that scales linearly and is highly accurate. FastOMA can process all >2000 eukaryotic UniProt ref proteomes <24 hours 🚀. Try it out at github.com/DessimozLab/fa…

From @SinaMajidian, @DessimozLab and colleagues, FastOMA achieves fast and accurate orthology inference, with linear scalability. nature.com/articles/s4159…

OMAnnotator: a novel approach to building an annotated consensus genome sequence doi.org/10.1101/2024.1… Automatic identification and annotation of MYB gene family members in plants doi.org/10.1101/2021.1…
In this exciting new preprint, Warwick Vesztrocy et al use Bgee to show that "the most diverged orthologues display the greatest change in gene expression, thus suggesting that they have a greater functional divergence" biorxiv.org/content/10.110…

Exploring gene duplications! Our study reveals how duplicated genes diverge in function using structural and expression analysis across 36 species. Check out our paper: biorxiv.org/content/10.110… Thanks to @the_alexwv, @Gloveface, Paul Thomas, @cdessimoz #Genomics #Evolution

Session 24: OMA/OMArk is starting now! To join in the session, jump over to our discord at: discord.gg/gF73fCF4 Or you can join in on the YouTube stream at: youtube.com/@thebiodiversi… @SinaMajidian @OMABrowser
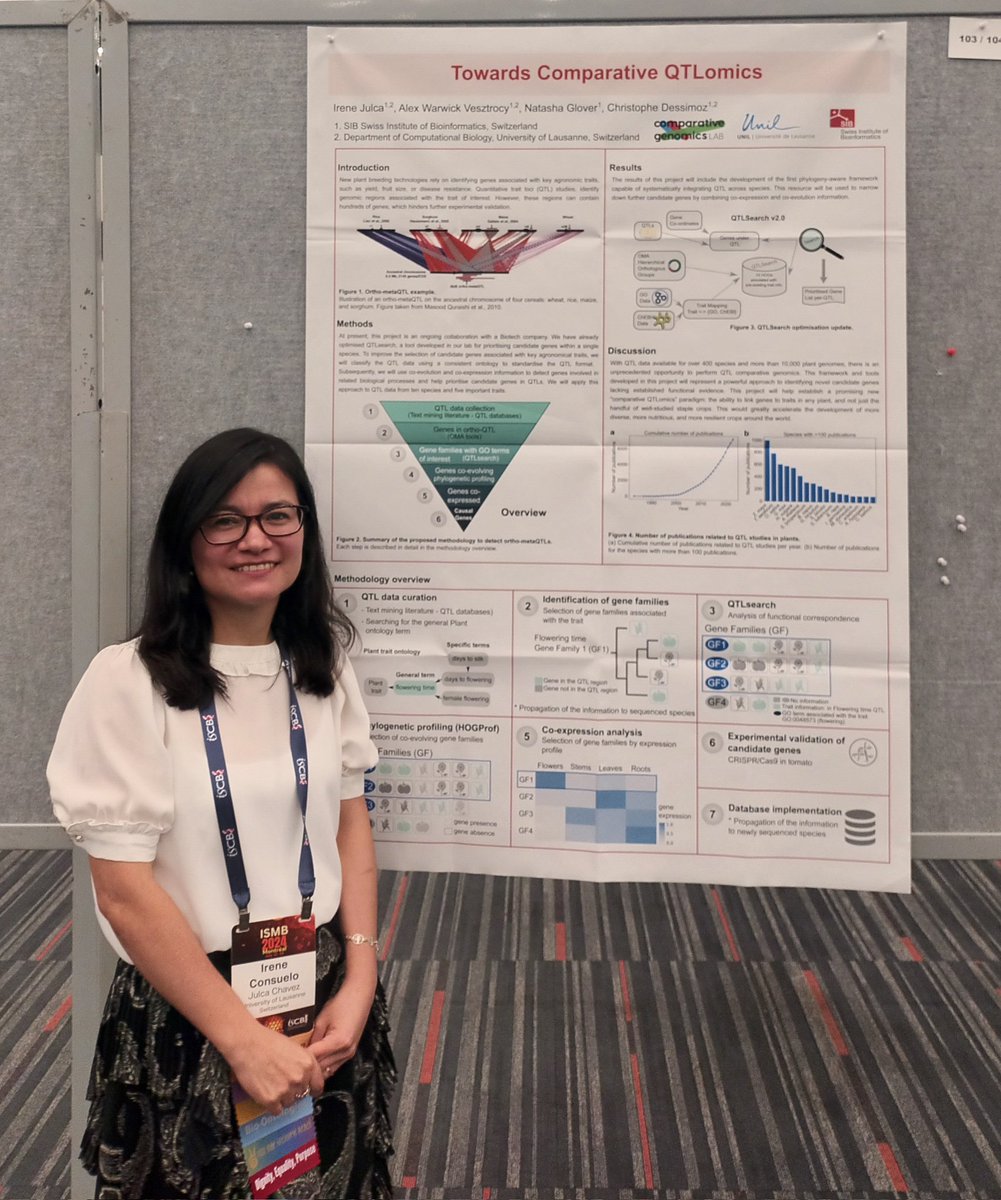
Many interesting discussions at the #ISMB2024 poster sessions. Thanks to everyone who stopped by our poster and to #ISCB and @DessimozLab for the opportunity!
With a nice photo of lab alumnus @leomrtns !
We made it! The final stop of @BioFAIRUK Roadshow. Lots of people to thank: -Those who has took the time to join us in Norwich -Our amazing speakers @irenepapatheodo @_EllenBell @DipaliD4 -Our fantastic colleagues @EarlhamInst for making the event run smoothly. #FAIR #DataSharing




nature.com/articles/s4155… Have a look at the article I reposted by @BallouxFrancois, @cedriccstan and @LucyvanDorp if you're interested in viruses and their evolutionary signature when jump hosts. I had the pleasure of writing a news and views piece about this work with @cdessimoz
Quality assessment of gene repertoire annotations with OMArk go.nature.com/4bH5OaF

📢 Glad to share our piece 'AI and the democratization of knowledge' @ScientificData. Data is the new oil—ripe for reuse. But what does it take to reuse? it's about making data interpretable by all, including AI, the ultimate non-specialist! #OpenScience nature.com/articles/s4159…
📢CALL FOR ABSTRACTS📢 🌟Abstract submission is now open for the Quest for Orthologs meeting #QFO8 which will be held at the University of Quebec in Montreal, Canada on July 17-18! More info at event.fourwaves.com/qfo8 Deadline: April 26.

How to evaluate the quality of a genome annotation? Introducing #OMArk, a groundbreaking tool by SIB’s @Why_Nevers & @Dessimozlab and part of the SIB Resource #SwissOrthology, now featured in @NatureBiotech: nature.com/articles/s4158…
Our OMArk paper is finally out at @NatureBiotech! I am excited to share the final results of our work with @Gloveface @cdessimoz @VictorRossier @DessimozLab. OMArk is a one-stop shop for eukaryotic genome annotation assessment. Link : nature.com/articles/s4158… (🧵)
United States Trends
- 1. Giannis N/A
- 2. Gigi N/A
- 3. Sinner N/A
- 4. Adam 22 N/A
- 5. #PersonaLive N/A
- 6. #SmackDown N/A
- 7. Trans N/A
- 8. #DragRace N/A
- 9. Bucks N/A
- 10. Jason Luv N/A
- 11. #JapanMegaExpo N/A
- 12. WILLIAMEST ECHO IN SG N/A
- 13. Pacers N/A
- 14. #WilliamEst1stFMinSingapore N/A
- 15. Jim Jones N/A
- 16. Antarctica N/A
- 17. Fritz N/A
- 18. Attitude Era N/A
- 19. Dabo N/A
- 20. Jen Z N/A
Something went wrong.
Something went wrong.