Krishnan Lab
@KrishnanLab
Laboratory for Biomedical Data Science and Computational Genomics at @CUBiomedInfo and @CUHealthAI | @[email protected]
You might like
📢#MLCB2024 Check out our recent review to break into translational biomedicine! We take a deep dive into methods for comparing & transferring data & knowledge across species, and highlight gaps/challenges. Details on implementation, data, & benchmarks: github.com/krishnanlab/cr…

Preprint 🚨 A review state-of-the-art computational strategies for cross-species knowledge transfer in biomedicine 💻👩🦰🐭🐟🪰🪱🧬🫁⚕️ Led by an excellent team at @KrishnanLab: @yhbioinfo, @ChrisAMancuso, & @kaylainbio in collab w/ @FishEvoDevoGeno 🧵 arxiv.org/abs/2408.08503
Preprint 🚨 A review state-of-the-art computational strategies for cross-species knowledge transfer in biomedicine 💻👩🦰🐭🐟🪰🪱🧬🫁⚕️ Led by an excellent team at @KrishnanLab: @yhbioinfo, @ChrisAMancuso, & @kaylainbio in collab w/ @FishEvoDevoGeno 🧵 arxiv.org/abs/2408.08503
1/ Excited to share Txt2onto 2.0, an approach combining language models & machine learning to annotate public samples & studies with standardized tissue & disease terms with a focus on interpretability & explainability! 📜 doi.org/10.1101/2024.0… 💻 github.com/krishnanlab/tx… 🧵👇🏼
Excited to see fruits of a huge community effort! Thanks to @ProfMilenkovic for the leadership and the opportunity to be a part of this 🙌🏽
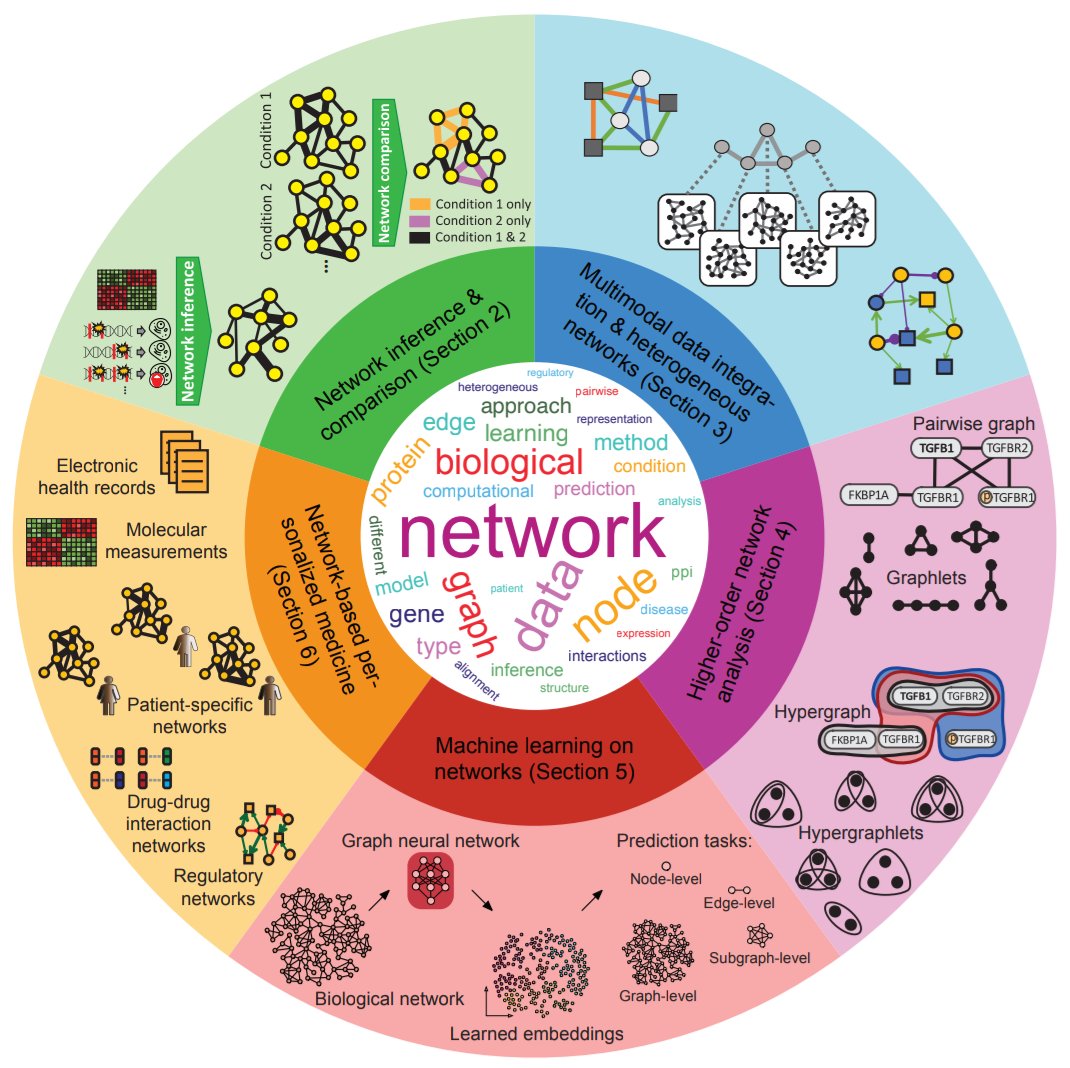
Our review+perspective paper on "Current and future directions in network biology" is online (arxiv.org/abs/2309.08478). This has been a Herculean effort by many co-authors, thank you all! Special thanks to @marinkazitnik, @_michellemli, @aydin_wells, and all section coordinators.
Excited to present new work led by @ChrisAMancuso: GenePlexusZoo, a computational framework to improve discovering new genes linked to pathways, phenotypes, & diseases both within & across species by combining molecular networks from multiple species w/ #machinelearning. [1/n]
Joint representation of molecular networks from multiple species improves gene classification biorxiv.org/cgi/content/sh… #bioRxiv
🎉CONGRATS to the winners of the @FASEBorg DataWorks! Prize🎉 🔁Grand Prize, Data Reuse: BrainChart — "Brain Charts for the Human Lifespan" 🤝Grand Prize, Data Sharing: N3C — "Democratizing Access to Clinical Data" Check out the winning projects: datascience.nih.gov/director/direc… #NIHData

Congrats to DataWorks! Challenge winners from our department! #N3C, led by @ontowonka, PhD, won the grand prize for #DataSharing, @MonarchInit, won the Distinguished Achievement Award for #Data Reuse, and @KrishnanLab PhD, won the Significant Achievement Award for Data Reuse!
🎉CONGRATS to the winners of the @FASEBorg DataWorks! Prize🎉 🔁Grand Prize, Data Reuse: BrainChart — "Brain Charts for the Human Lifespan" 🤝Grand Prize, Data Sharing: N3C — "Democratizing Access to Clinical Data" Check out the winning projects: datascience.nih.gov/director/direc… #NIHData

The Ramanathan Lab is looking for a Research Tech to join us! We are a young and exciting group interested in how cells of diverse shapes and sizes form functional tissue. Please DM if you are interested in the position or want to know more! unmc.peopleadmin.com/postings/76294 Please RT!
Three @CUAnschutz teams are DataWorks prize finalists! Learn more about @KrishnanLab, @MonarchInit, and #NationalCovidCohortCollaborative's projects and vote for one here👇
Learn about the 20 DataWorks! Prize finalists and how they have used data sharing or reuse to advance human health. Finalists are eligible for $500,000 prize purse and people’s choice recognition awards. #NIHData #DataScience herox.com/dataworks
A new software created by @compbiologist, PhD, associate professor of biomedical informatics at @CUMedicalSchool, and his team, annotates genomics samples based on unstructured sample text descriptions, making them easier for researchers to find and use.👇 nature.com/articles/s4146…
The @KrishnanLab develops an approach combining natural language processing and machine learning to infer the source tissue of public genomics samples based on their plain text descriptions, making these samples easy to discover and reuse @compbiologist go.nature.com/3UIi9lE
nature.com
Systematic tissue annotations of genomics samples by modeling unstructured metadata
Nature Communications - The 1+ million publicly-available human –omics samples currently remain acutely underused. Here the authors present an approach combining natural language processing...
This wk, @kaylainbio defended her brilliant PhD work on using #transcriptomes, #networks, & #machinelearning to probe age- & sex-specificity 🎉 Being the first grad student from @KrishnanLab, Kayla has set a v. high bar! Congratulations Dr. Johnson! We're very proud of you!!




🎇 We're one of 20 finalists for the DataWorks! Prize for our work on: Removing metadata barriers to promote data reuse Methods that address unstructured & missing metadata that are barriers to discovering & reusing public omics data. Learn more & VOTE! herox.com/dataworks/roun…
The @KrishnanLab develops an approach combining natural language processing and machine learning to infer the source tissue of public genomics samples based on their plain text descriptions, making these samples easy to discover and reuse @compbiologist go.nature.com/3UIi9lE
nature.com
Systematic tissue annotations of genomics samples by modeling unstructured metadata
Nature Communications - The 1+ million publicly-available human –omics samples currently remain acutely underused. Here the authors present an approach combining natural language processing...
Automated NLP and ML approach to annotate samples to their tissue-of-origin by modeling unstructured metadata. Improved performance and scalability compared to MetaSRA @NatureComms @compbiologist nature.com/articles/s4146…
The Computational Biosciences PhD Program @CUAnschutz is holding an Info Session on Fri, Oct. 21 for interested students to get to know the program! Plus, all registered attendees will receive an application fee waiver!! Please spread the word. graduateschool.cuanschutz.edu/events-and-res…
Two posters in @MlcsbC #ISMB2022 @NeutralYh | W-058 Cross-species transcriptome-based regression to discover model equivalents of human samples & genes. w/ @ingo_braasch @filjev46 | W-067 Determining the optimal embedding technique for mapping transcriptomes to ontologies
We have a bunch of cool projects to share @ #ISMB2022! #TextMining #NetBio @OBF_BOSC @TransMedISMB @MlcsbC #BioVis Interested in working with? Chat with our team! They are great people and you can also learn about how it is to work with us.

We have 3 posters at @TransMedISMB #ISMB2022 @kaylainbio | N-023 Age- and sex-specific gene signatures and networks @slepphickey | N-024 Chronic inflammation network signatures in complex diseases @KewalinSamart | Virtual Drug repurposing for infectious diseases w/ @janani137
We have a bunch of cool projects to share @ #ISMB2022! #TextMining #NetBio @OBF_BOSC @TransMedISMB @MlcsbC #BioVis Interested in working with? Chat with our team! They are great people and you can also learn about how it is to work with us.

We have 2 posters at #NetBio #ISMB2022 @ChrisAMancuso | L-016 Jointly modeling networks from multiple species to improve gene classification Alex McKim | L-017 Module-based prediction improves network-based prioritization of genes associated w/ complex traits & diseases
We have a bunch of cool projects to share @ #ISMB2022! #TextMining #NetBio @OBF_BOSC @TransMedISMB @MlcsbC #BioVis Interested in working with? Chat with our team! They are great people and you can also learn about how it is to work with us.

On Wed, July 13, @ChrisAMancuso is giving a talk in @OBF_BOSC #BOSC on "GenePlexus: A web server and Python package for gene discovery using network-based machine learning" Paper: doi.org/10.1093/nar/gk… Webserver: geneplexus.net Repo: github.com/krishnanlab/ge… #ISMB2022
github.com
GitHub - krishnanlab/geneplexus-app-old
Contribute to krishnanlab/geneplexus-app-old development by creating an account on GitHub.
We have a bunch of cool projects to share @ #ISMB2022! #TextMining #NetBio @OBF_BOSC @TransMedISMB @MlcsbC #BioVis Interested in working with? Chat with our team! They are great people and you can also learn about how it is to work with us.

United States Trends
- 1. Good Monday 27.1K posts
- 2. TOP CALL 3,691 posts
- 3. AI Alert 1,381 posts
- 4. #centralwOrldXmasXFreenBecky 540K posts
- 5. SAROCHA REBECCA DISNEY AT CTW 560K posts
- 6. #BaddiesUSA 65.8K posts
- 7. #MondayMotivation 7,364 posts
- 8. #LingOrmDiorAmbassador 287K posts
- 9. NAMJOON 54.3K posts
- 10. Market Focus 2,511 posts
- 11. Check Analyze N/A
- 12. Token Signal 1,826 posts
- 13. Rams 30.3K posts
- 14. #LAShortnSweet 24.1K posts
- 15. DOGE 181K posts
- 16. Chip Kelly 9,242 posts
- 17. Vin Diesel 1,561 posts
- 18. Scotty 10.4K posts
- 19. Stacey 23.6K posts
- 20. Gilligan 4,046 posts
Something went wrong.
Something went wrong.