Robin H. van der Weide
@RobinWeide
Postdoc Integrative Single-Cell Functional Genomics in the @kind_jop lab (@_Hubrecht). PhD from @NKI_nl
You might like
Happy to see the improved and expanded version of this out in @naturemethods 🎉 nature.com/articles/s4159…
📯 Thread alert: introducing MAbID, a new combinatorial single-cell profiling method. This was a team effort between PhD-candidate Silke Lochs and me, with the full support of the @kind_jop lab at @_Hubrecht. Read all about it: biorxiv.org/content/10.110…

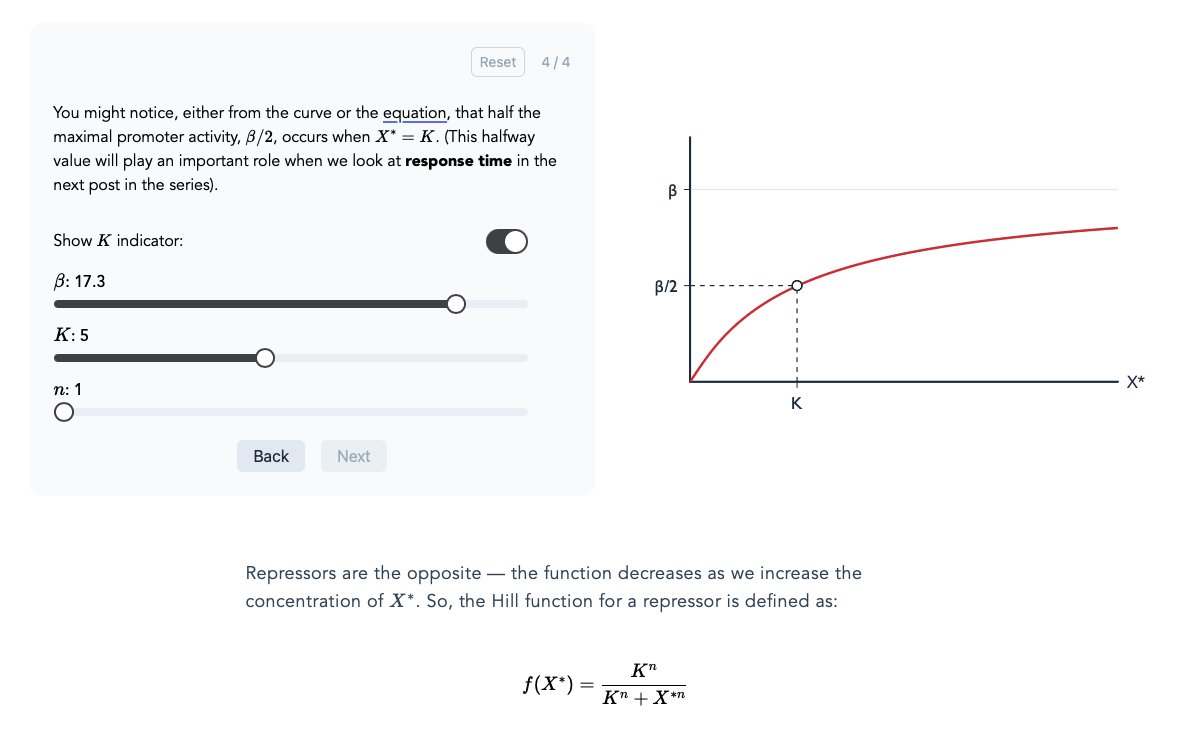
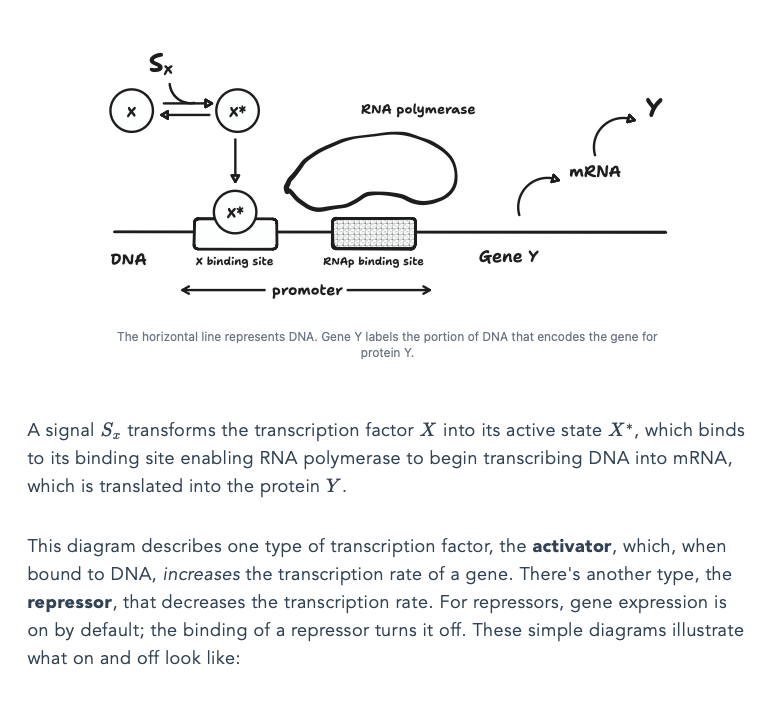
.@nehaludyavar is doing interesting work. He's currently reading Uri Alon's book to learn Systems Biology. As he learns about new concepts, he writes about them and makes super clean graphics + interactives to accompany the text. He's "learning in public" in a beautiful way.
So thrilled to see our work on regulation of transcriptional repression in lamina-associated published in @embojournal. Thanks to all the authors!! Thanks to all the authors for their fantastic contribution. embopress.org/doi/full/10.10…
Great to see @AleksSzczurek's paper out today! A wonderful team effort with @emmy_dimitrova , @Jess_R_Kelley , and @blackledge_neil. Have a look if you are interested in how #Polycomb controls transcription to repress gene expression. rdcu.be/dTD3t
4) The scientific method also applies to computational results: proper controls and negating all alternative hypotheses are just as important in computational biology as in bench experiments.
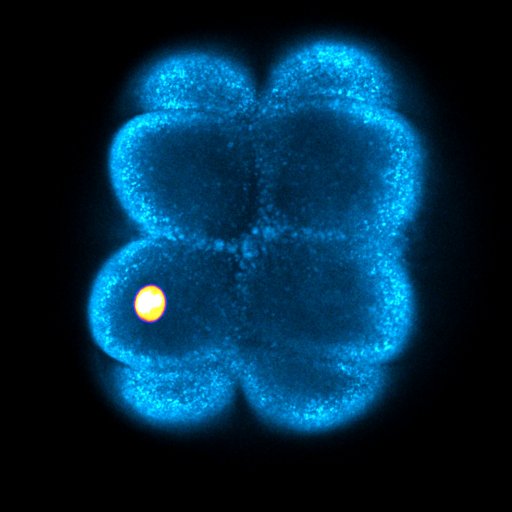
Out today in @NatureMaterials, the updated version of our work on nuclear jamming in organogenesis 👁️🧠 Thanks to the reviewers (yes, thanks 😊), we have added a lot of new data. Check below for the new results... 🧵 ➡️rdcu.be/dQDm6

How do enhancers work over distances that sometimes exceed megabases? Excited to share our work led by @gracecbower where we uncover a unique sequence signature globally associated with long-range enhancer-promoter interactions in developing limb buds: biorxiv.org/content/10.110… 1/

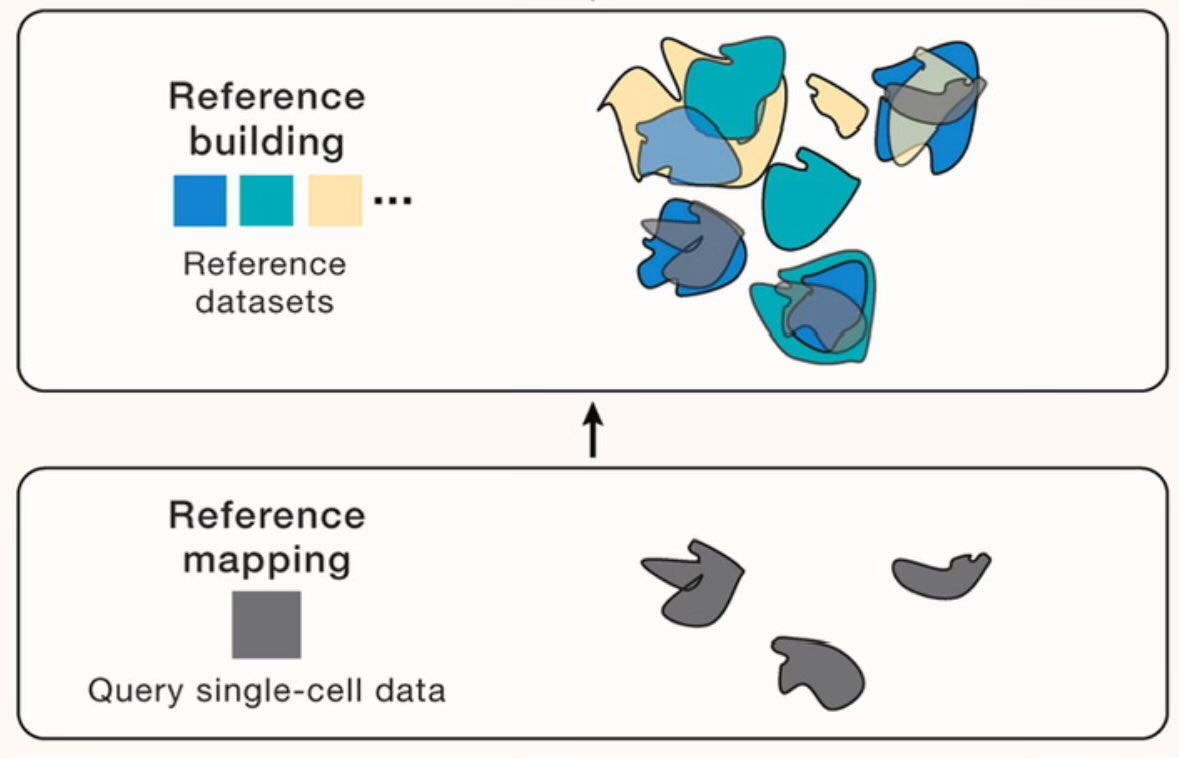
In need of an educational weekend read? Check out our new perspective @CellCellPress with Rahul @satijalab. Led by @mo_lotfollahi & @YUHANHAO2, we outline how to automate single-cell data analysis via reference mapping. cell.com/cell/fulltext/…
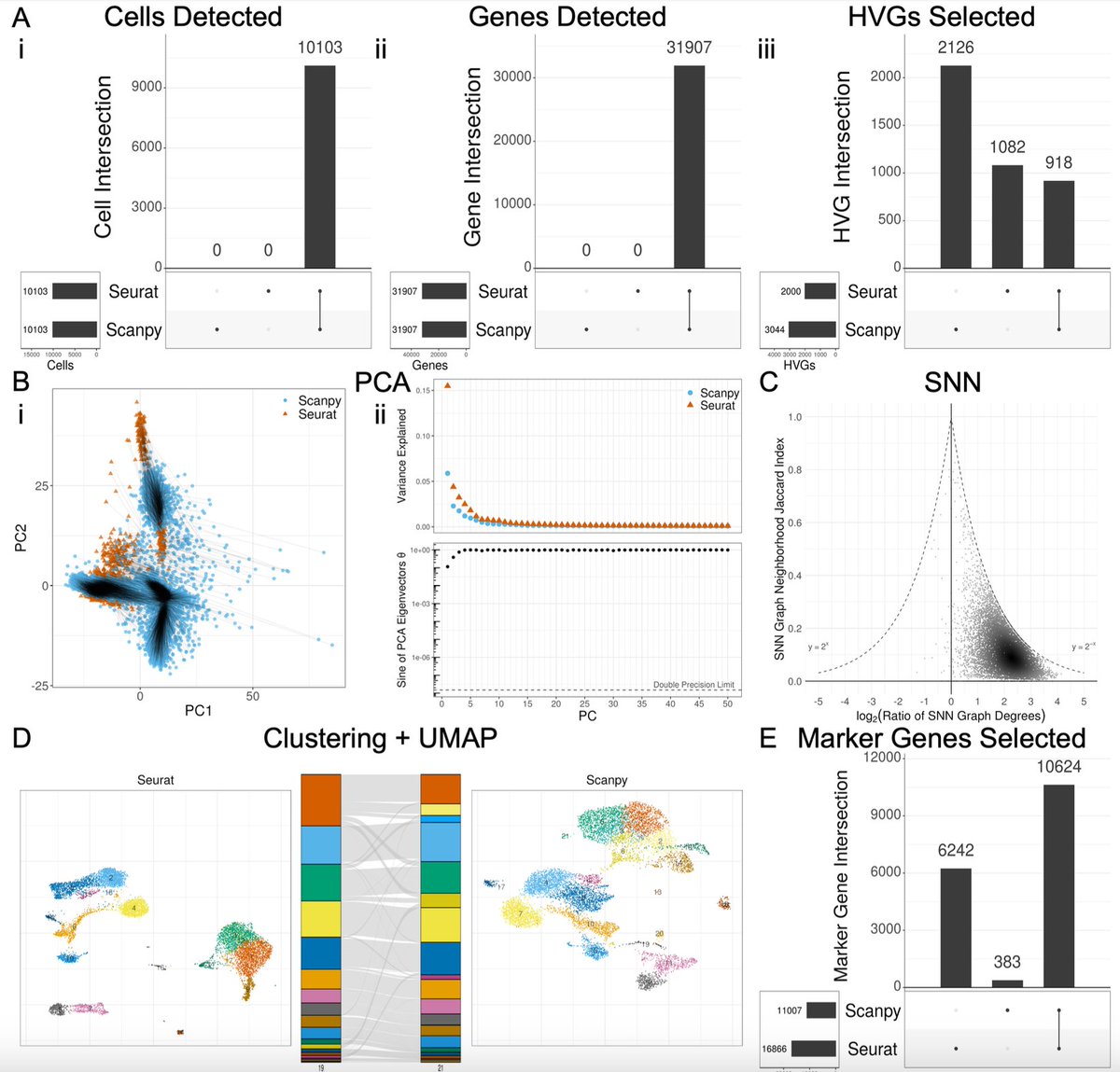
The choice of whether to use Seurat or Scanpy for single-cell RNA-seq analysis typically comes down to a preference of R vs. Python. But do they produce the same results? In biorxiv.org/content/10.110… w/ @Josephmrich et al. we take a close look. The results are 👀 1/🧵
In this preprint biorxiv.org/content/10.110… with @sindri_e we compared seven widely used methods for batch correction of single cell RNA-seq data. We found that all but one of the methods introduce batch effects when there are none. 1/N
(1/14) Excited to share our new work on ZNF143/ZFP143 by @DomenicNarducci ZNF143 has been widely reported to be a general loop factor Instead we and @deWitLab in a companion preprint show that ZNF143 is a TF with no loop function: biorxiv.org/content/10.110… biorxiv.org/content/10.110…

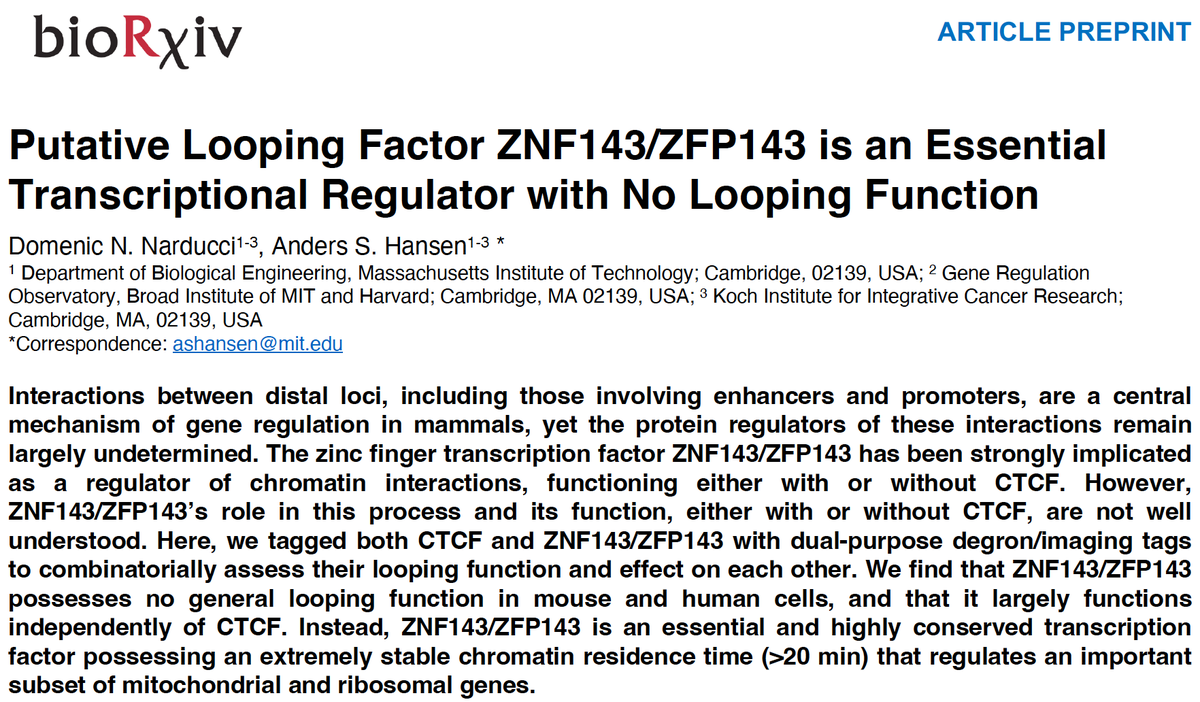
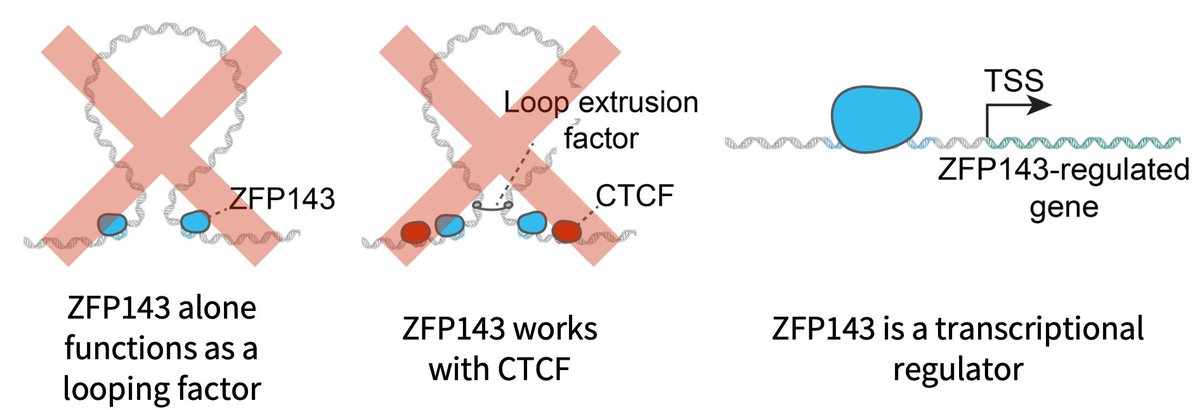
You may know ZNF143 as a looping factor. We show together with the @hansen_lab that this is not the case! If you want to know why and what it does instead do read our thread below: biorxiv.org/content/10.110…
Congratulations to Puck Knipscheer and @kind_jop, who have been awarded @NWOFunding Vici grants for their research on resolving dangerous DNA structures and the role of chromosome organization in determining cell identity respectively.

We have an open post-doc position in the lab. If you're interested in studying the role of the cohesin complex in gene regulation, don't hesitate to apply. Come and join us in beautiful Amsterdam! Email me if you have specific questions. nki.nl/careers-study/…
Last January, I noticed something peculiar in my 2yo’s bedroom that - after a year of obsessive reporting - led me to a profound cosmic revelation about what’s even possible in our universe. A 🧵.

📢We are very excited to announce the second edition of the Dutch Embryo Model Meeting #DEMM24 Are you working with embryo models such as 2D/3D #gastruloids or #blastoids? Join our meeting and submit an abstract! oncodeinstitute.nl/events/dutch-e… @oncodeinstitute

New preprint from our lab: LAD scrambling to figure out how they interact with the nuclear lamina. biorxiv.org/content/10.110…
Next week is the 42nd time my lab will run the journal club (JC) in a lab meeting in the 'collaborative' format, which we do 1/month, and I think it is the ultimate journal-club format, so a 🧵 about it
Exciting work by @kind_jop lab published in @naturemethods! Read this news item to learn more about MAbID: ru.nl/onderzoek/onde… ru.nl/en/research/re…
The Kind lab @_Hubrecht describe MAbID, a multiplexing approach to uncover the genomic distributions of various epigenetic markers. They demonstrate joint measurements of six epitopes in mouse bone marrow at single-cell resolution. @kind_jop @RobinWeide nature.com/articles/s4159…
United States Trends
- 1. Ryan Garcia N/A
- 2. #UFCHouston N/A
- 3. Strickland N/A
- 4. Rondale Moore N/A
- 5. Rockets N/A
- 6. Knicks N/A
- 7. Duke N/A
- 8. Brunson N/A
- 9. Auburn N/A
- 10. Greenland N/A
- 11. UCLA N/A
- 12. Fluffy N/A
- 13. Michigan N/A
- 14. Mark Pope N/A
- 15. Chandler N/A
- 16. Jose Alvarado N/A
- 17. Frank Martin N/A
- 18. #AEWCollision N/A
- 19. Albright N/A
- 20. Hiraoka N/A
Something went wrong.
Something went wrong.