#geneticcodeexpansion search results
Congratulations to @kosuke_seki on completing a terrific PhD. So amazed at his creative innovations in #synbio and #geneticcodeexpansion @NUSynBio @NorthwesternEng . Kosuke is headed to @KortemmeLab.

Check out our new paper in @angew_chem describing an engineered tRNA that initiates protein synthesis with non-canonical amino acids IN VIVO! #GeneticCodeExpansion #synbio @cgem_cci @AlannaSchepartz @jhdcate @amikura1986 @Fredrward @YaleChem @YaleMBB onlinelibrary.wiley.com/doi/10.1002/an…

New minireview just accepted in @ChemBioChem! Here we discuss strategies for initiating protein synthesis with diverse molecules in vitro and in vivo. @YaleMBB @Yale @cgem_cci #SynBio #GeneticCodeExpansion onlinelibrary.wiley.com/doi/abs/10.100…

Check out our new paper in @NatureComms! We describe a novel method for #PhageDisplay with an expanded genetic code and the development of potent inhibitors of the histone deacetylase SIRT2. #GeneticCodeExpansion @TAMUScience @TAMUChemistry @liulabtamu1 tinyurl.com/r589wrz

Programmed #ubiquitin #acetylation using #GeneticCodeExpansion reveals altered #ubiquitination patterns →febs.onlinelibrary.wiley.com/doi/10.1002/18…

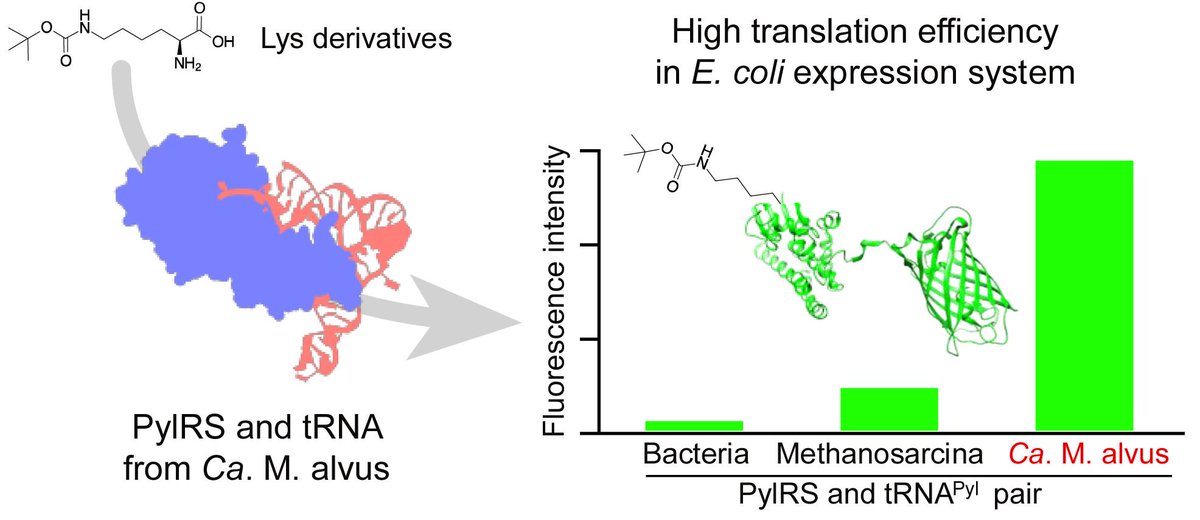
Pyrrolysyl-tRNA Synthetase with a Unique Architecture Enhances the Availability of Lysine Derivatives in Synthetic Genetic Codes sci.fo/5iu #GeneticCodeExpansion #Archaea #ProteinEngineering @Molecules_MDPI
Study by @theshawlab @westernuBiochem highlighted in our new issue: Programmed #ubiquitin #acetylation using #geneticcodeexpansion reveals altered #ubiquitination patterns →febs.onlinelibrary.wiley.com/doi/10.1002/18…

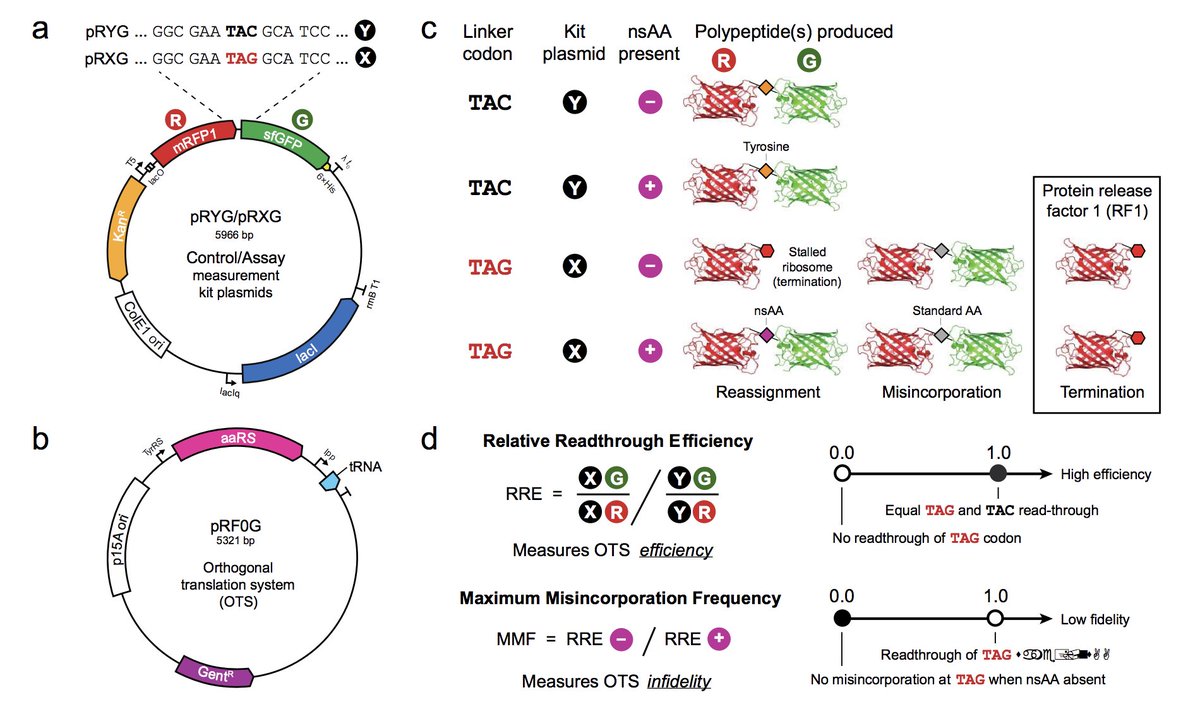
New plasmids from @barricklab. Rapid and inexpensive evaluation of nonstandard amino acid incorporation in Escherichia coli, published in @ACSSynBio. #geneticcodeexpansion #synbio hubs.ly/H0ddHgR0
#mdpimolecules Recent Development of Genetic Code Expansion for Posttranslational Modification Studies mdpi.com/313686 #GeneticCodeExpansion #PosttranslationalModification #SyntheticBiology @UArkansas
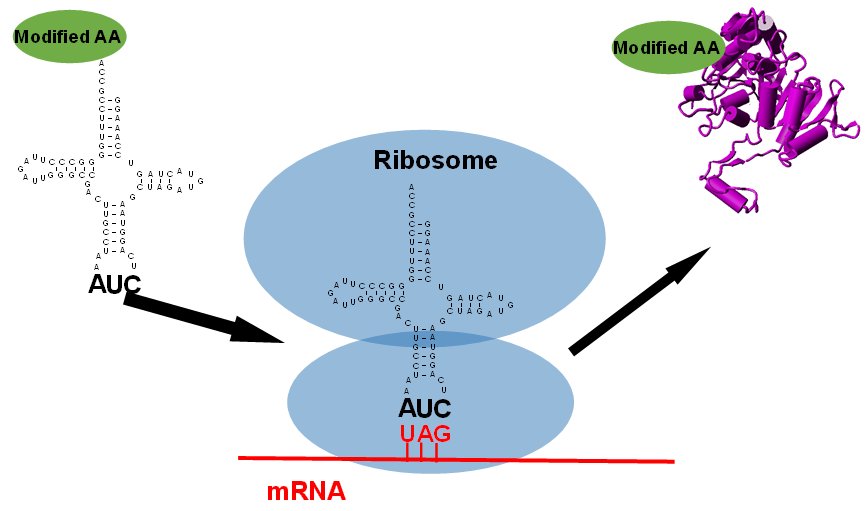
Our new preprint is live @bioRxiv! We used an engineered initiator tRNA to initiate protein synthesis (with noncanonical amino acids) at a reassigned sense codon. Coupling this with nonsense suppression enable genetic encoding of 3 distinct ncAAs. #synbio #GeneticCodeExpansion
Genetic Encoding of Three Distinct Noncanonical Amino Acids Using Reprogrammed Initiator and Nonsense Codons biorxiv.org/cgi/content/sh… #bioRxiv
Great work from the Marchand lab (marchandlab.com)! They enzymatically linked non-standard bases to standard ones, and they developed a nanopore-based platform to identify such new bases through sequencing 👌🧬. #synbio #geneticcodeexpansion nature.com/articles/s4146…
Emerging trends in applying in vivo #geneticcodeexpansion strategies for selection, addiction and #catalysis (Mayer @univgroningen) #ChemBioTalents doi.wiley.com/10.1002/cbic.2…

Our review on new in cellulo applications for unnatural amino acids is out #geneticcodeexpansion Congrats Kristina! 🥳🙌 Link provides 50 days Open Access authors.elsevier.com/a/1biOi4sz6M8W…
Embark on a non-natural amino acid journey! 🌟 Read about the exciting story of expanding the genetic code. 🧬 doi.org/10.1042/bio_20… #GeneticCodeExpansion #AminoAcidResearch #ScienceStory

Today in @PNASNews we describe the engineering of split aminoacyl-tRNA synthetases for proximity-induced stop codon suppression. This was a very fun project spearheaded by two super talented students! @IUSM_Biochem @YaleMBB #SynBio #GeneticCodeExpansion pnas.org/doi/10.1073/pn…
Pairing intracellular synthesis of noncanonical amino acids with #GeneticCodeExpansion to foster biotechnological applications (Biava @BrevardCollege) doi.wiley.com/10.1002/cbic.2…

PhD in Enzyme Engineering with Genetic Code Expansion 📍 University of Zurich, Switzerland Apply now: jobs.uzh.ch/job-vacancies/… #PhDpositions #EnzymeEngineering #GeneticCodeExpansion #UniversityOfZurich #ScienceCareers
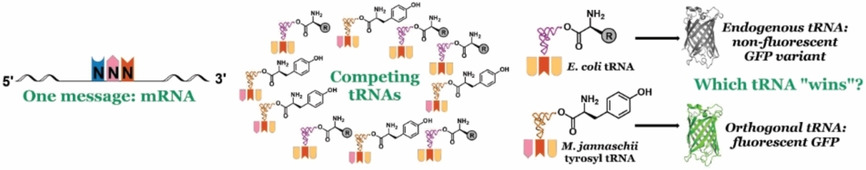
The influence of competing tRNA abundance on translation: Quantifying the efficiency of sense codon reassignment at rarely used codons #syntheticbiology #geneticcodeexpansion #proteinengineering (Fisk @CUSystem @CUDenver) doi.wiley.com/10.1002/cbic.2…
This video by @SciShow does a great job explaining #GeneticCodeExpansion for a lay audience; however, I totally disagree with the video title. The genetic code is amazing, and optimized in ways that we have yet to fully understand. youtube.com/watch?v=9j7oEu…

youtube.com
YouTube
How We Could Rewrite Our Genetic Code
PhD in Enzyme Engineering with Genetic Code Expansion 📍 University of Zurich, Switzerland Apply now: jobs.uzh.ch/job-vacancies/… #PhDpositions #EnzymeEngineering #GeneticCodeExpansion #UniversityOfZurich #ScienceCareers
Our platform simplifies workflows in protein engineering and chemical biology. Huge thanks to @ANUChemistry @ARC_CIPPS @arc_gov_au for their support! 📖 Read more: doi.org/10.1016/j.chem… #BioorthogonalChemistry #GeneticCodeExpansion #ProteinEngineering #ChemicalBiology 2/2
#TPbiotecUV 👉Teaching enzymes new reactions through #GeneticCodeExpansion and #DirectedEvolution: chemistryworld.com/news/teaching-…
4/ Opuu and Simonson (@Polytechnique) review the application of computational design methods to noncanonical amino acids for #GeneticCodeExpansion. academic.oup.com/peds/article/d…
Engineering stress response pathways can increase the efficiency of genetic code expansion in mammalian cells @LemkeLab @uni_mainz @uni_mainz_eng @imbmainz. #GeneticCodeExpansion, #ProteinEngineering, #synbio. #BiotechNatureComms nature.com/articles/s4146…
nature.com
Remodeling the cellular stress response for enhanced genetic code expansion in mammalian cells
Nature Communications - Genetic code expansion (GCE) is a protein engineering tool that enables programmed and site-specific installation of noncanonical amino acids into proteins. Here, authors...
Great work from the Marchand lab (marchandlab.com)! They enzymatically linked non-standard bases to standard ones, and they developed a nanopore-based platform to identify such new bases through sequencing 👌🧬. #synbio #geneticcodeexpansion nature.com/articles/s4146…
Embark on a non-natural amino acid journey! 🌟 Read about the exciting story of expanding the genetic code. 🧬 doi.org/10.1042/bio_20… #GeneticCodeExpansion #AminoAcidResearch #ScienceStory

Congratulations to @kosuke_seki on completing a terrific PhD. So amazed at his creative innovations in #synbio and #geneticcodeexpansion @NUSynBio @NorthwesternEng . Kosuke is headed to @KortemmeLab.

🧬🔬Our latest research paper showcases GCEXpress, an optimized GCE system, as a powerful tool to study the adhesive, mechanical, and signaling properties of aGPCRs and their ligand interactions. Don't miss out! onlinelibrary.wiley.com/doi/10.1002/pr… #GPCR #GeneticCodeExpansion #ClickChemistry
Today in @PNASNews we describe the engineering of split aminoacyl-tRNA synthetases for proximity-induced stop codon suppression. This was a very fun project spearheaded by two super talented students! @IUSM_Biochem @YaleMBB #SynBio #GeneticCodeExpansion pnas.org/doi/10.1073/pn…
#OpenAccess Selective Inhibition of Cysteine-Dependent Enzymes by Bioorthogonal Tethering buff.ly/3eSldMy @UoDLifeSciences @mrcppu @SatpalVirdee , Dr Yu-Hsuan Tsai et al. #GeneticCodeExpansion #Chemogenetics
Congrats the the @Mehl_lab @RyanAMehl1 on an awesome paper! #geneticcodeexpansion
Check out our latest paper in @ScienceAdvances where we create designer tetrazine-based anti SARS-Cov nanobody conjugations with very interesting results! When it comes to nanobody conjugation, control is key. #nanobody #chembio science.org/doi/10.1126/sc…
Check out the news on the new NIH Funded GCE4ALL Research Center @OSUScience @Mehl_lab @RyanAMehl1 #GCE4ALL #geneticcodeexpansion kval.com/news/local/osu…
Using a quadruplet codon to expand the genetic code of an animal. #GeneticCodeExpansion academic.oup.com/nar/advance-ar… @NAR_Open
This video by @SciShow does a great job explaining #GeneticCodeExpansion for a lay audience; however, I totally disagree with the video title. The genetic code is amazing, and optimized in ways that we have yet to fully understand. youtube.com/watch?v=9j7oEu…

youtube.com
YouTube
How We Could Rewrite Our Genetic Code
Another big 'first' in C. elegans! Congratulations to the @GreissLab team. #GeneticCodeExpansion #Celegans #synbio
Using a Quadruplet Codon to Expand the Genetic Code of an Animal biorxiv.org/cgi/content/sh… #bioRxiv
Some theoretical aspects of reprogramming the standard genetic code. #GeneticCodeExpansion academic.oup.com/genetics/advan… #Genetics @GeneticsGSA
Great talks today at the ADSE Young Researchers Conference! I’m excited to share our work on #GeneticCodeExpansion tomorrow afternoon.
Our new preprint is live @bioRxiv! We used an engineered initiator tRNA to initiate protein synthesis (with noncanonical amino acids) at a reassigned sense codon. Coupling this with nonsense suppression enable genetic encoding of 3 distinct ncAAs. #synbio #GeneticCodeExpansion
Genetic Encoding of Three Distinct Noncanonical Amino Acids Using Reprogrammed Initiator and Nonsense Codons biorxiv.org/cgi/content/sh… #bioRxiv
Our review on new in cellulo applications for unnatural amino acids is out #geneticcodeexpansion Congrats Kristina! 🥳🙌 Link provides 50 days Open Access authors.elsevier.com/a/1biOi4sz6M8W…
Congratulations to @kosuke_seki on completing a terrific PhD. So amazed at his creative innovations in #synbio and #geneticcodeexpansion @NUSynBio @NorthwesternEng . Kosuke is headed to @KortemmeLab.

Study by @theshawlab @westernuBiochem highlighted in our new issue: Programmed #ubiquitin #acetylation using #geneticcodeexpansion reveals altered #ubiquitination patterns →febs.onlinelibrary.wiley.com/doi/10.1002/18…

Programmed #ubiquitin #acetylation using #GeneticCodeExpansion reveals altered #ubiquitination patterns →febs.onlinelibrary.wiley.com/doi/10.1002/18…

Pyrrolysyl-tRNA Synthetase with a Unique Architecture Enhances the Availability of Lysine Derivatives in Synthetic Genetic Codes sci.fo/5iu #GeneticCodeExpansion #Archaea #ProteinEngineering @Molecules_MDPI

Check out our new paper in @angew_chem describing an engineered tRNA that initiates protein synthesis with non-canonical amino acids IN VIVO! #GeneticCodeExpansion #synbio @cgem_cci @AlannaSchepartz @jhdcate @amikura1986 @Fredrward @YaleChem @YaleMBB onlinelibrary.wiley.com/doi/10.1002/an…

#mdpimolecules Recent Development of Genetic Code Expansion for Posttranslational Modification Studies mdpi.com/313686 #GeneticCodeExpansion #PosttranslationalModification #SyntheticBiology @UArkansas

New minireview just accepted in @ChemBioChem! Here we discuss strategies for initiating protein synthesis with diverse molecules in vitro and in vivo. @YaleMBB @Yale @cgem_cci #SynBio #GeneticCodeExpansion onlinelibrary.wiley.com/doi/abs/10.100…

Check out our new paper in @NatureComms! We describe a novel method for #PhageDisplay with an expanded genetic code and the development of potent inhibitors of the histone deacetylase SIRT2. #GeneticCodeExpansion @TAMUScience @TAMUChemistry @liulabtamu1 tinyurl.com/r589wrz

Embark on a non-natural amino acid journey! 🌟 Read about the exciting story of expanding the genetic code. 🧬 doi.org/10.1042/bio_20… #GeneticCodeExpansion #AminoAcidResearch #ScienceStory

Emerging trends in applying in vivo #geneticcodeexpansion strategies for selection, addiction and #catalysis (Mayer @univgroningen) #ChemBioTalents doi.wiley.com/10.1002/cbic.2…

New plasmids from @barricklab. Rapid and inexpensive evaluation of nonstandard amino acid incorporation in Escherichia coli, published in @ACSSynBio. #geneticcodeexpansion #synbio hubs.ly/H0ddHgR0

Pairing intracellular synthesis of noncanonical amino acids with #GeneticCodeExpansion to foster biotechnological applications (Biava @BrevardCollege) doi.wiley.com/10.1002/cbic.2…

The influence of competing tRNA abundance on translation: Quantifying the efficiency of sense codon reassignment at rarely used codons #syntheticbiology #geneticcodeexpansion #proteinengineering (Fisk @CUSystem @CUDenver) doi.wiley.com/10.1002/cbic.2…

Something went wrong.
Something went wrong.
United States Trends
- 1. GeForce Season 4,184 posts
- 2. Comey 192K posts
- 3. Everton 147K posts
- 4. Mark Kelly 127K posts
- 5. St. John 8,763 posts
- 6. Amorim 62.7K posts
- 7. #sjubb N/A
- 8. Iowa State 3,662 posts
- 9. UCMJ 19K posts
- 10. Seton Hall 2,431 posts
- 11. 49ers 20.3K posts
- 12. Manchester United 84.8K posts
- 13. Genesis Mission 2,698 posts
- 14. Opus 4.5 9,544 posts
- 15. Dealing 31K posts
- 16. Hegseth 51K posts
- 17. #LightningStrikes N/A
- 18. Pickford 11.5K posts
- 19. Benedict Arnold 4,230 posts
- 20. Noesen N/A