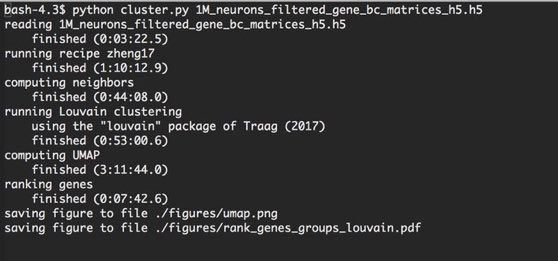
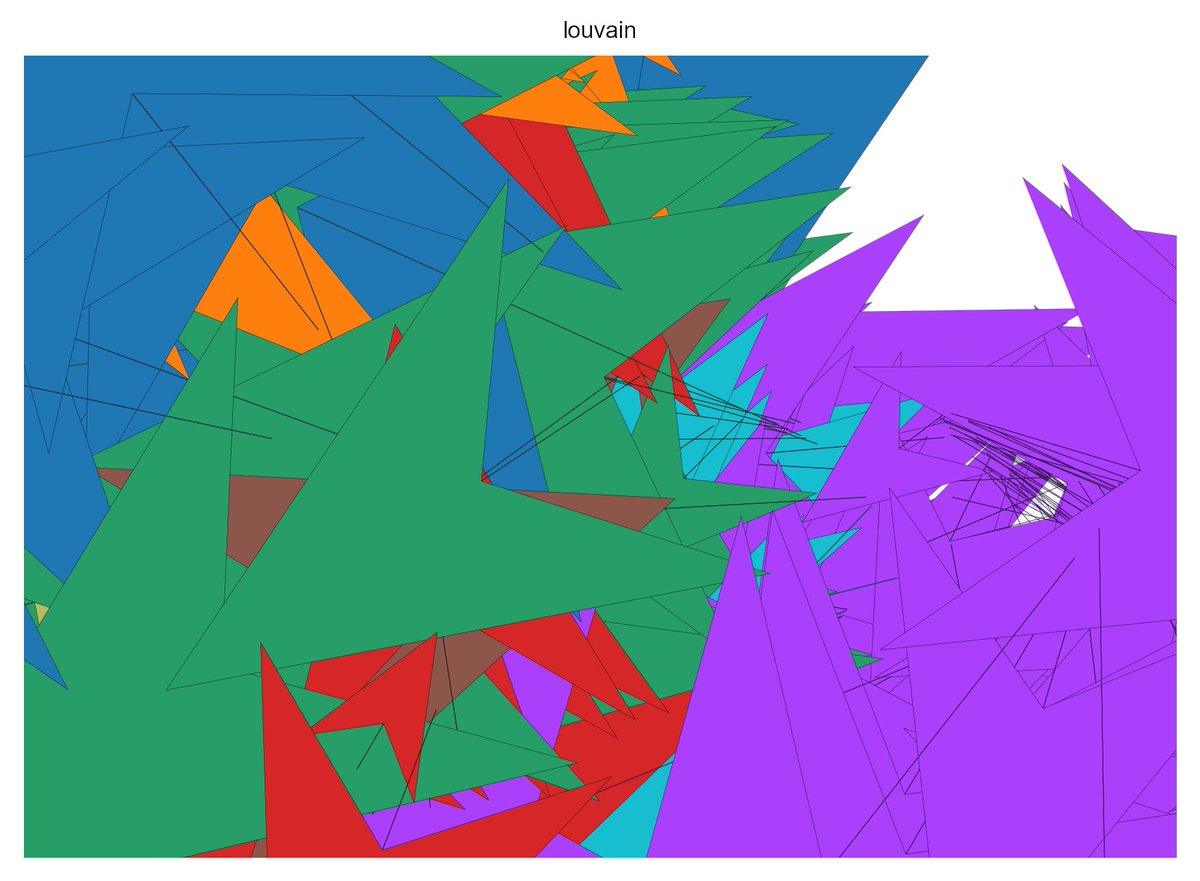
#scanpy نتائج البحث
#scanpy can also analyse bulk RNA-seq data 😎 Partition-based graph abstraction (PAGA) and UMAP results on 11k #GTEx samples are really cool.
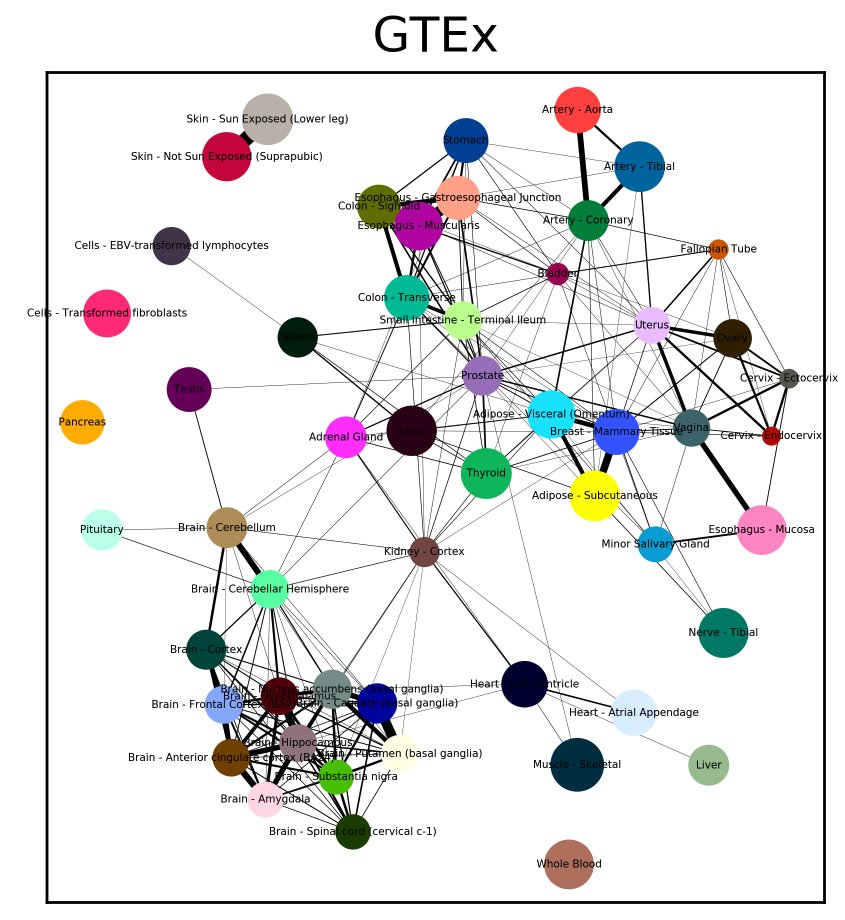
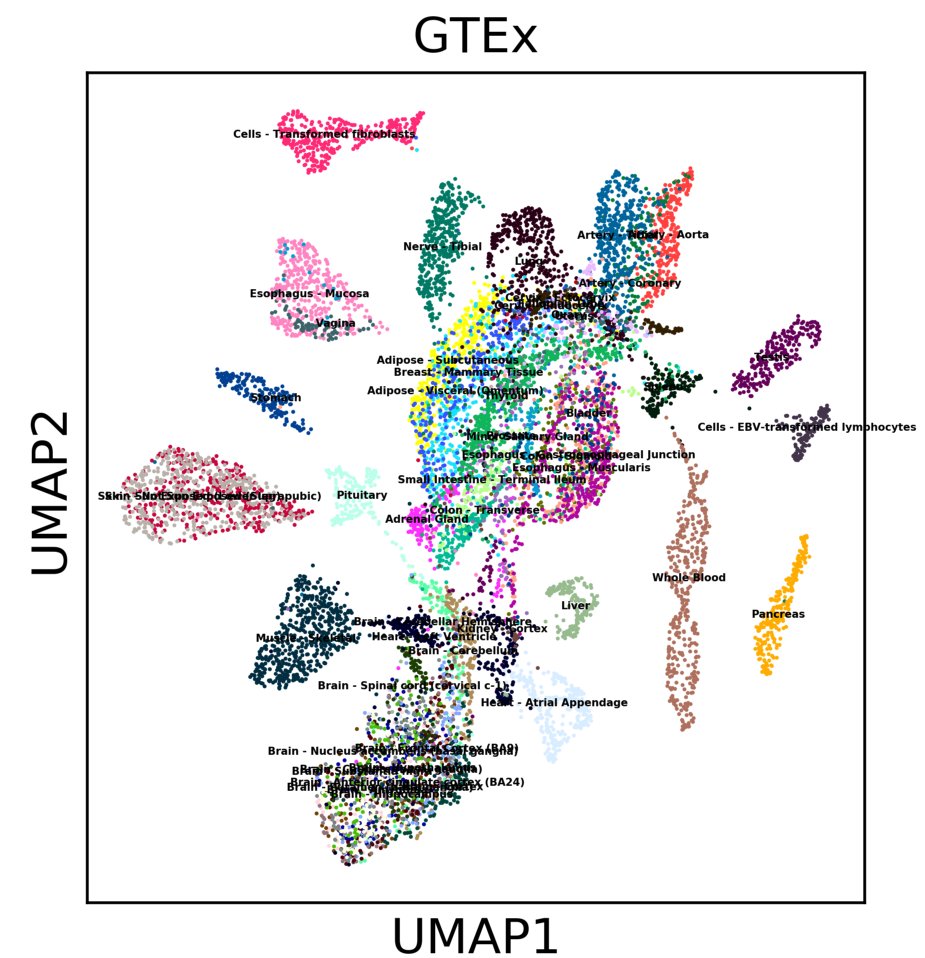
We added UMAP support to scanpy today! It seems to work very well on single cell data. Another reason to try #scanpy #SingleCell
The series "UMAP beasts and where to find them" continues. This time I spotted a shrimp 🦐, which is funny as data were processed using #scanpy (check the project's logo) #UMAPbeasts
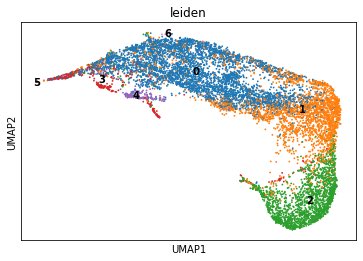
Awesome demo of the upcoming @humancellatlas data portal by @gen_haliburton & happy to see her and the team using #scanpy for this
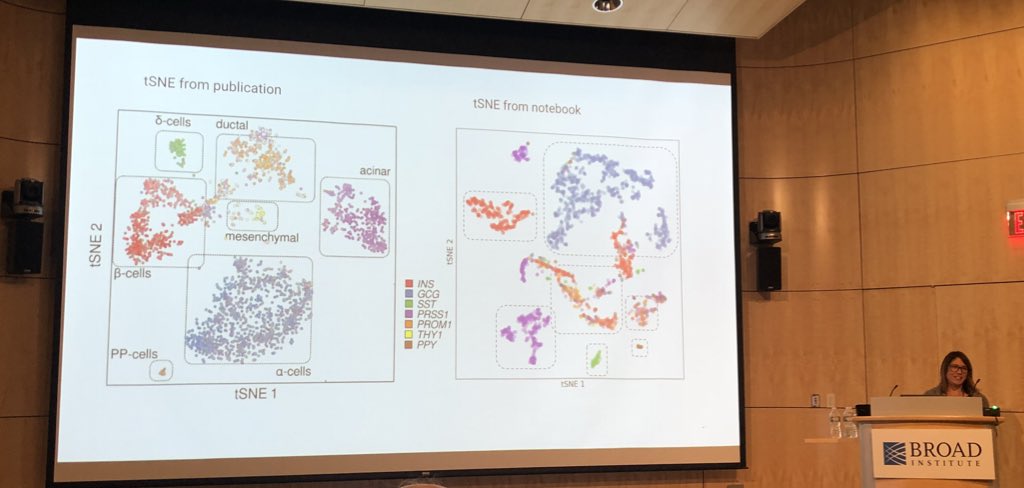
Excited to share 'Squidpy', our toolbox for spatial single cell analysis. Led by @g_palla1 & @HannahSpitzer1, we extend #Scanpy's infrastructure and analyses to store, manipulate & interactively visualize spatial omics data. Read at biorxiv.org/content/10.110… squidpy.readthedocs.io
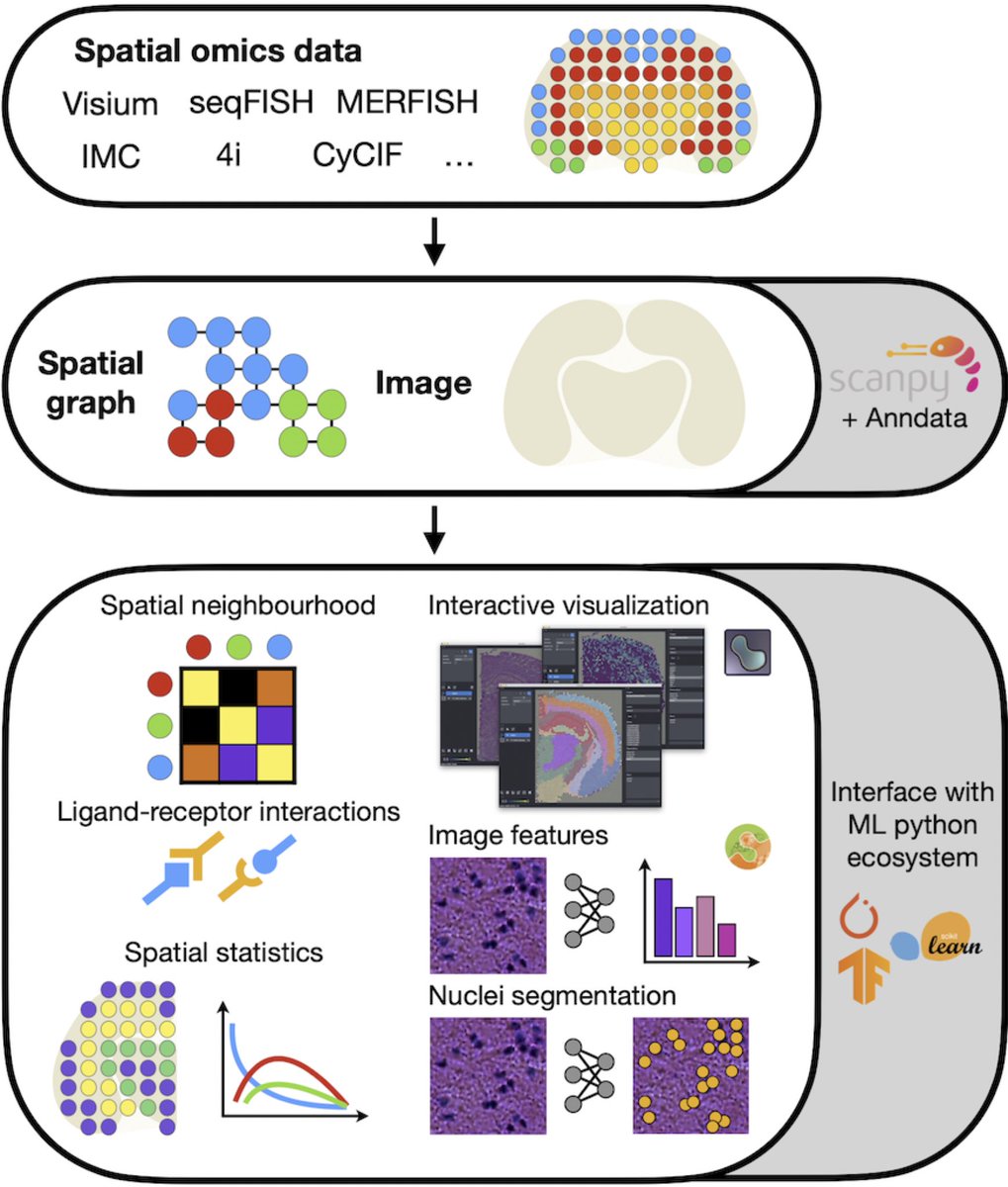
Scarf is very memory-efficient when compared against two awesome single-cell tools, Seurat and Scanpy. No runtime overhead. With Scarf, one can analyze atlas scale datasets from their laptop or run several of them in parallel on server. #Seurat #Scanpy #scRNAseq 2/8
A workshop on #singlecell RNA-seq data analysis using #scanpy was held at Cellular and Molecular Research Center, Birjand University of Medical Sciences, from 22 to 27 August 2020.



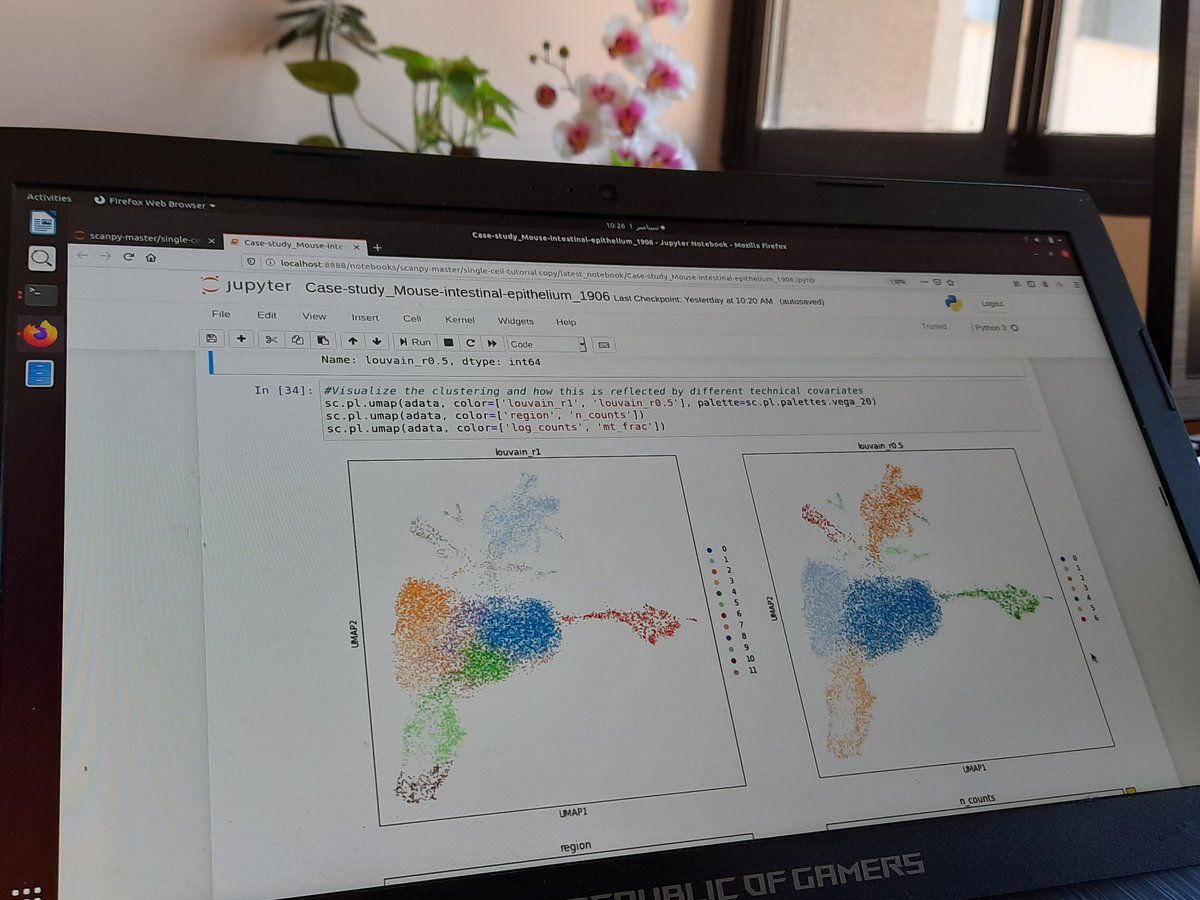
Single cell proteomics finally becoming a reality, with awesome miniaturization effort by @labs_mann! Robust plate-based readout (>1200 proteins), and strong differences to RNA; we (@_sabrinarichter @davidsebfischer ) contributed adapted #scanpy analyses. biorxiv.org/content/10.110…

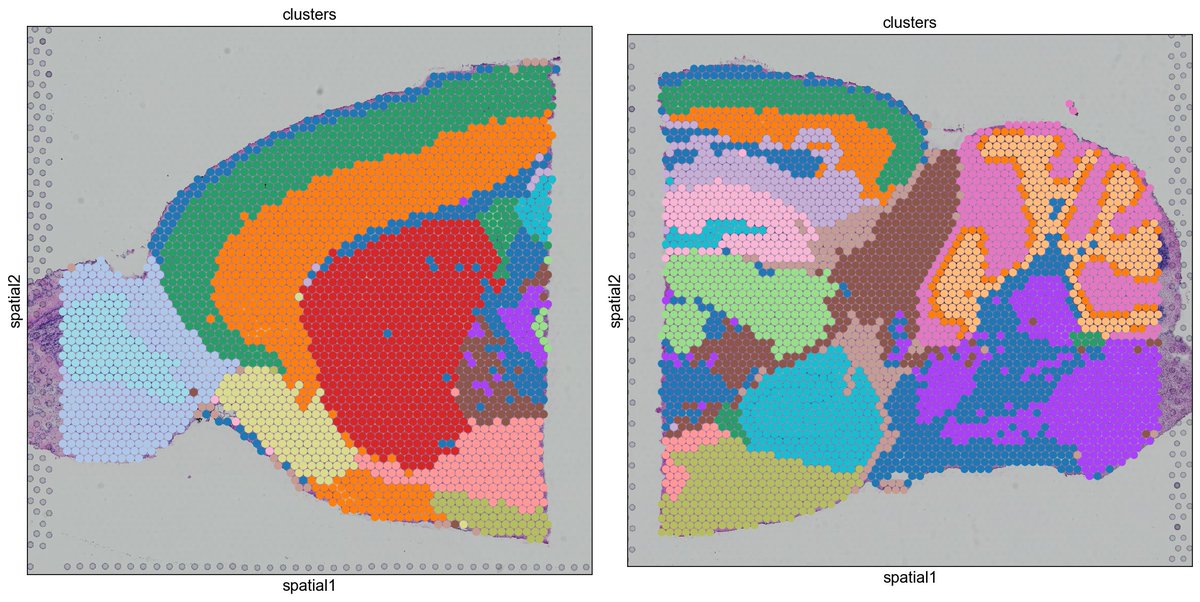
We're excited to release a #scanpy update for the analysis and visualization of #spatialTranscriptomics data, focussing on @10xGenomics Visium and MERFISH, thx @g_palla1. Tutorial at nbviewer.jupyter.org/github/theisla… Support for different data types and integration with #scRNAseq soon!

Really happy how well the new #scanpy stickers turned out - happy to share, eg at next conferences (Hinxton, Florence). 🤓

New #Scanpy and anndata releases are out, thanks to all the great contributors! New features include robust backend support for multiple slices in and data integration in spatial scRNA-seq, see tutorials: scanpy-tutorials.readthedocs.io/en/latest/spat… scanpy-tutorials.readthedocs.io/en/latest/spat…
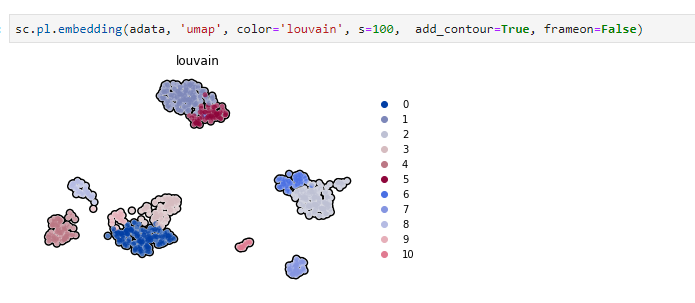
Just made a PR to #scanpy to produce these type of plots. I call them contour for the time being but I would like to find a better name. Does anyone know how these scatter plots with borders are called?
Collaboration in #scRNA data analysis doesn’t have to be challenging. Explore how C-DIAM can help by letting bioinformatics teams upload and share outputs from #Seurat or #Scanpy with bench scientists for interactive viz. Read our blog for more details: pythiabio.com/post/share-and…
Experience the magic of Tahoe-100M thanks to @fabian_theis's group and #Scanpy.
Interested in exploring @vevo_ai’s massive Tahoe-100M single-cell perturbation dataset? #Scanpy now supports out-of-core analysis, powered by Dask—allowing you to seamlessly work with a single huge AnnData object without loading everything into memory. 🔗 theislab.github.io/vevo_Tahoe_100…
Interested in exploring @vevo_ai’s massive Tahoe-100M single-cell perturbation dataset? #Scanpy now supports out-of-core analysis, powered by Dask—allowing you to seamlessly work with a single huge AnnData object without loading everything into memory. 🔗 theislab.github.io/vevo_Tahoe_100…
Hopefully this should solve the fight between #Seurat and #scanpy : easySCF: A Tool for Enhancing Interoperability Between R and Python for Efficient Single-Cell Data Analysis | Bioinformatics | Oxford Academic academic.oup.com/bioinformatics…
Chrysalis can be easily integrated into any workflow based on #Scanpy and works seamlessly with multi-sample datasets. A huge thanks again to Jelica Vasiljevic, Kerstin Hahn, @RottenbergSven, and @alvaldeolivas for their invaluable contributions. 🧵 (6/7)
scATAcat is available as a Python package (github.com/aybugealtay/sc…), compatible with #scanpy & #anndata, and comes with a tutorial available at scatacat.readthedocs.io/en/latest/inde…
💯 Absolutely loved this comprehensive and insightful comparison on #Seurat vs #Scanpy for #scRNAseq analysis
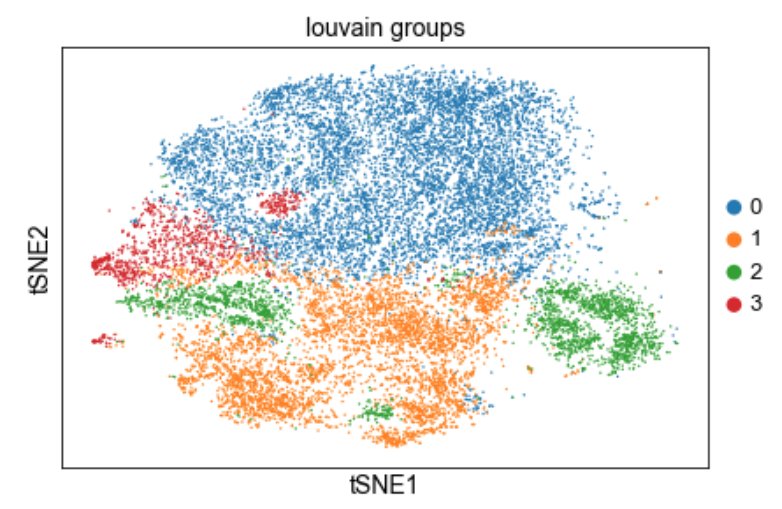
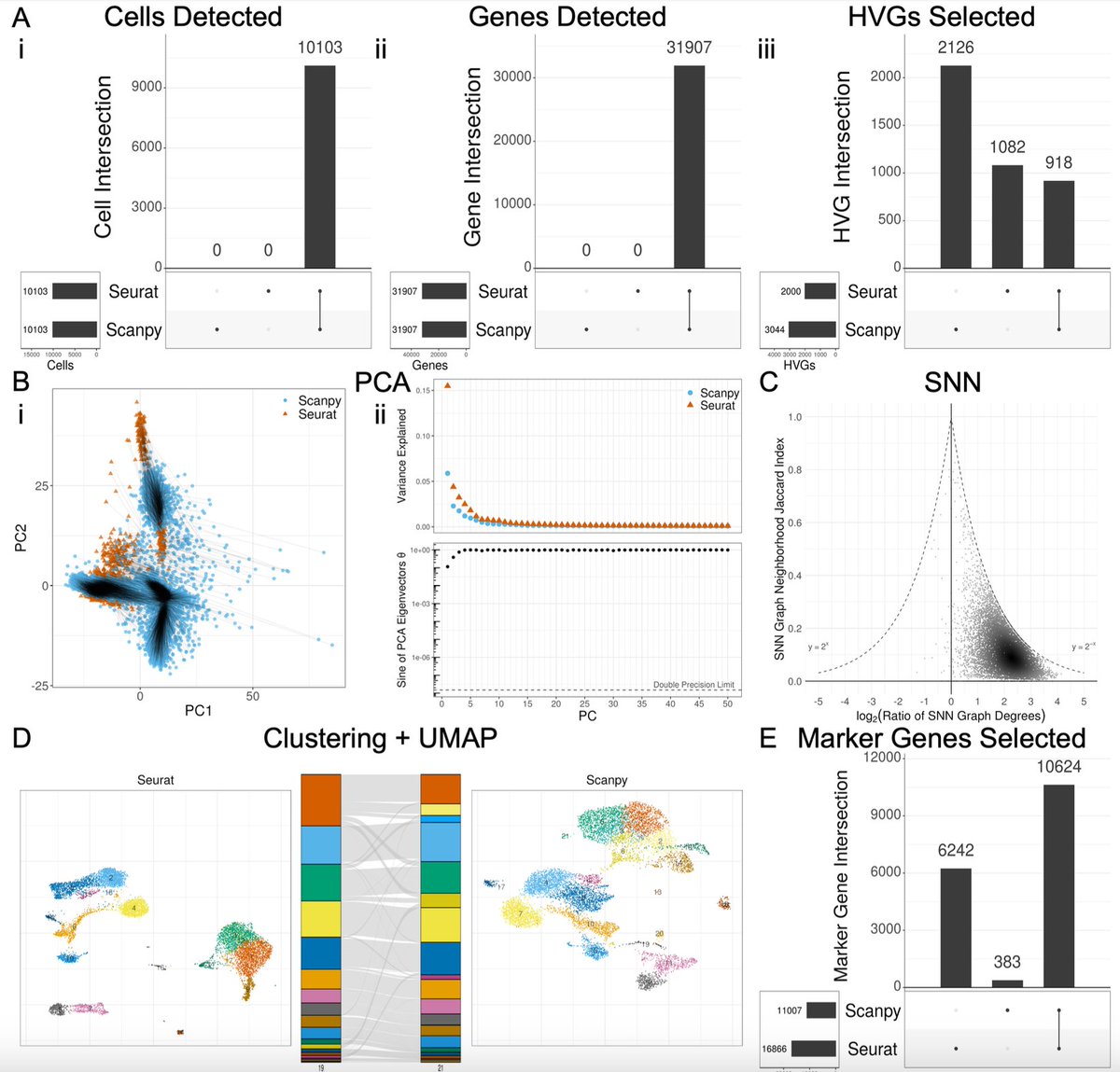
The choice of whether to use Seurat or Scanpy for single-cell RNA-seq analysis typically comes down to a preference of R vs. Python. But do they produce the same results? In biorxiv.org/content/10.110… w/ @Josephmrich et al. we take a close look. The results are 👀 1/🧵
Probably one of the most comprehensive scRNA seq popular softwares ( @satijalab #seurat & @scverse_team #scanpy ) most comprehensive comparison. Loved it.
The choice of whether to use Seurat or Scanpy for single-cell RNA-seq analysis typically comes down to a preference of R vs. Python. But do they produce the same results? In biorxiv.org/content/10.110… w/ @Josephmrich et al. we take a close look. The results are 👀 1/🧵

If you are doing single cell RNA sequencing, you must have thoughts on Trajectory analysis. There are multiple tools on the github, but which one you used and why ? #seurat #scanpy #Bioinformatics #scRNA
Chrysalis can be easily integrated into any workflow based on #scanpy and works on multi-sample datasets. Huge thanks to Jelica Vasiljevic, Kerstin Hahn, @RottenbergSven, and @alvaldeolivas for their invaluable contributions. 🧵 (5/6)
Authors have used @scverse_team #scanpy for gene expression pre-processing , Episcanpy for chromatin accessibility ,Batch integration and scMultiomics integration was performed using scib.metrices ! /2
This package extends the Infinity Flow protocol to allow for more complex configurations. Data is held in AnnData Python objects to enable seamless integration with popular single-cell omics tools, such as #scanpy and #pytometry. (3/7)
This package extends the Infinity Flow protocol to allow for more complex configurations. Data is held in AnnData Python objects to enable seamless integration with popular single-cell omics tools, such as #scanpy and #pytometry. (2/6)
I've been exploring #GPT models' capabilities in #singlecell analysis for a while now, and they've all struggled with #scanpy. Although #ChatGPT-3.5 showed promise, it fell short in grasping the complexities of single cell RNA-seq data. Enter #GPT4. 👇
Introducing #CellTypeWriter, a user-friendly tool to explore and analyze #scRNAseq data using an interactive chat interface powered by #GPT4. Check it out on github: github.com/ntranoslab/cel…
#scanpy can also analyse bulk RNA-seq data 😎 Partition-based graph abstraction (PAGA) and UMAP results on 11k #GTEx samples are really cool.


We added UMAP support to scanpy today! It seems to work very well on single cell data. Another reason to try #scanpy #SingleCell

The series "UMAP beasts and where to find them" continues. This time I spotted a shrimp 🦐, which is funny as data were processed using #scanpy (check the project's logo) #UMAPbeasts

We're excited to release a #scanpy update for the analysis and visualization of #spatialTranscriptomics data, focussing on @10xGenomics Visium and MERFISH, thx @g_palla1. Tutorial at nbviewer.jupyter.org/github/theisla… Support for different data types and integration with #scRNAseq soon!

Awesome demo of the upcoming @humancellatlas data portal by @gen_haliburton & happy to see her and the team using #scanpy for this

Excited to share 'Squidpy', our toolbox for spatial single cell analysis. Led by @g_palla1 & @HannahSpitzer1, we extend #Scanpy's infrastructure and analyses to store, manipulate & interactively visualize spatial omics data. Read at biorxiv.org/content/10.110… squidpy.readthedocs.io

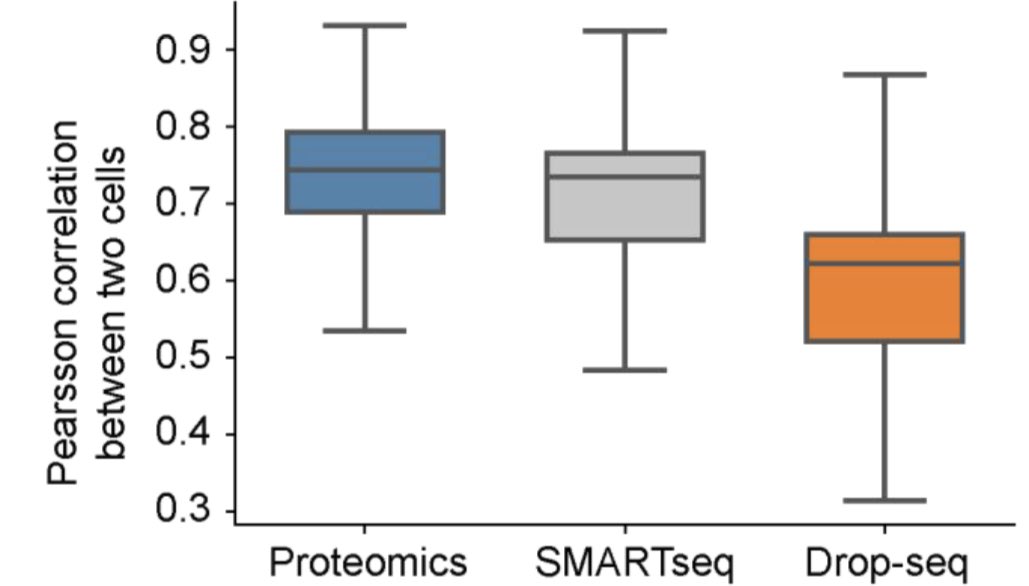
Single cell proteomics finally becoming a reality, with awesome miniaturization effort by @labs_mann! Robust plate-based readout (>1200 proteins), and strong differences to RNA; we (@_sabrinarichter @davidsebfischer ) contributed adapted #scanpy analyses. biorxiv.org/content/10.110…


A workshop on #singlecell RNA-seq data analysis using #scanpy was held at Cellular and Molecular Research Center, Birjand University of Medical Sciences, from 22 to 27 August 2020.




Really happy how well the new #scanpy stickers turned out - happy to share, eg at next conferences (Hinxton, Florence). 🤓

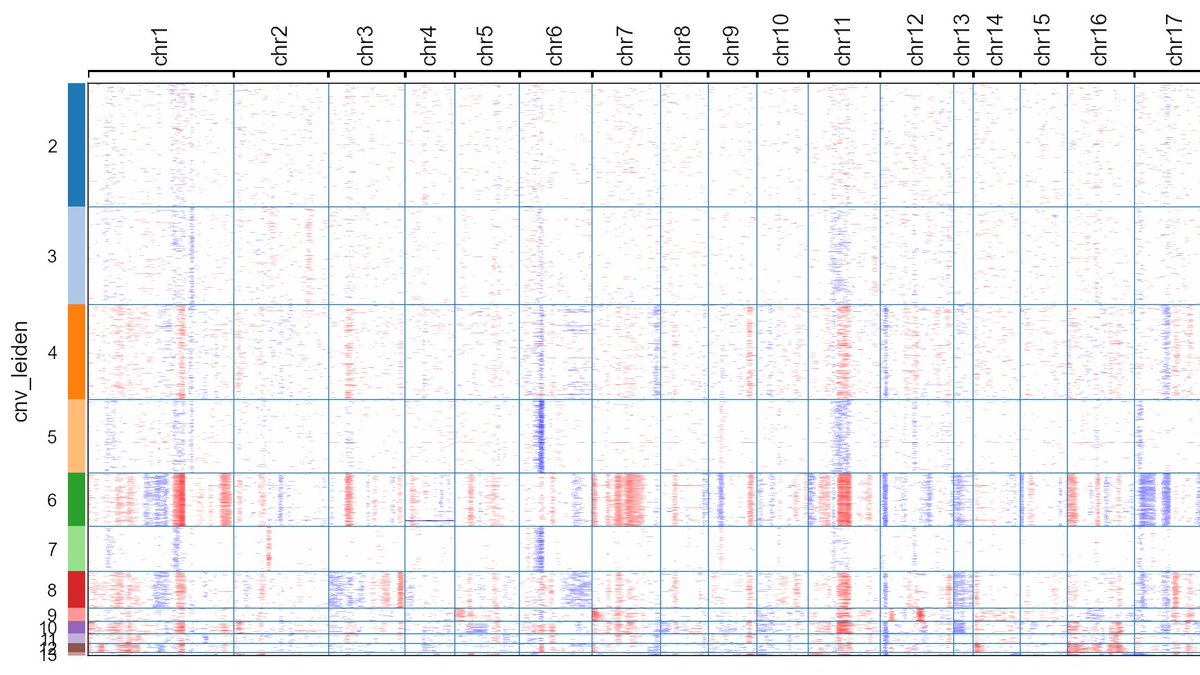
I created inferCNVpy, a Python re-implementation of @broadinstitute's inferCNV. Works smoothly with #scanpy and is ~100x faster than the R version. Already applied it to 500k cells. github.com/icbi-lab/infer… (1/3)
Something went wrong.
Something went wrong.
United States Trends
- 1. Brunson 7,085 posts
- 2. Knicks 13.2K posts
- 3. Derek Dixon N/A
- 4. Jaylen Brown 7,006 posts
- 5. Celtics 14.3K posts
- 6. Mark Pope N/A
- 7. Notre Dame 36.5K posts
- 8. Duke 29K posts
- 9. Caleb Wilson N/A
- 10. Jordan Walsh 1,003 posts
- 11. Miami 97.1K posts
- 12. Braylon Mullins N/A
- 13. Bama 23.7K posts
- 14. Van Epps 129K posts
- 15. #WWENXT 11.3K posts
- 16. Kentucky 27.6K posts
- 17. Tennessee 218K posts
- 18. #kubball 1,122 posts
- 19. Nashville 33.5K posts
- 20. Mikal Bridges 1,584 posts