#spatialtranscriptomic 搜尋結果
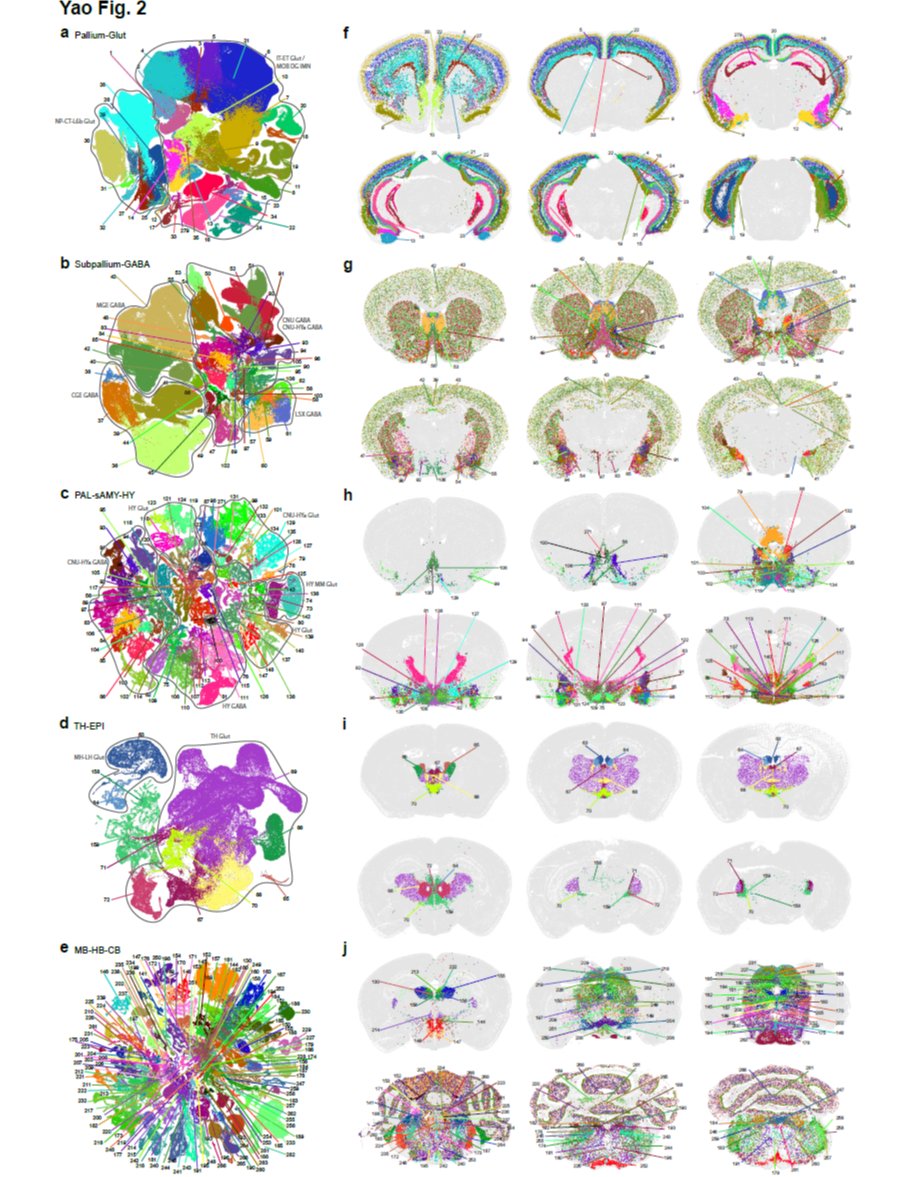
A🐭🧠 project spearheaded by @AllenInstitute created a #SpatialTranscriptomic #CellTypeAtlas of the whole #MouseBrain! Many key marker genes used in our new #MERSCOPE PanNeuro Cell Type Panel were validated in this project. Read the paper & learn more: hubs.ly/Q01KGWK90
Thrilled to receive the NIH/NIGMS R35 MIRA to support our #bioinformatics work studying subcellular and cellular #spatialtranscriptomic heterogeneity Special thanks to @rajivmccoy for guidance + support from mentors @jtleek, Joel Bader, Mike Miller @JHUBME 🥳🥂 #AcademicChatter

Starting today....@nanostringtech Nanostring GEOMx fully operational and training beginning! @UofGMVLS @pvanhouts Mathias Holpert @UofGCancerSci @CRUKGlasgow looking forward to exciting #spatialtranscriptomic projects @CRUK_BI @grisurgery @hollyleslie_ @EdinCRC @precisionpanc




@OliverStegle presenting on identification of spatially variable genes #spatialtranscriptomic #cytodata2019

STMiner, an algorithm specifically designed for complex #spatialtranscriptomic data can remove false-positive genes from the analysis results. We are deeply appreciative of @CellGenomics for affording us the platform to present this significant research outcome.

1/4 #SpatialTranscriptomic methods allow to analyse the gene expression of tissues without losing its spatial heterogeneity. In my #PhD in the Meier lab @PioneerCampus I apply and advance such methods to address different biological questions. #YoungScientistsHMGU

Welcome to Tu Duyen Nguyen, @AllianceProg intern from @Polytechnique. Tu Duyen is joining the IICD to focus on novel algorithms to understand the spatial interaction between cells from #spatialtranscriptomic data in the @elhamazizi lab.

Check out 🔻🔻 #cholangiocarcinoma Paper from @chiarabraconi @ColmJJORourke gut.bmj.com/content/early/… Well done @LabSpatialNBJ for #spatialtranscriptomic component @Gut_BMJ @grisurgery @nanostringtech @UofGMVLS @UofGCancerSci


@mason_lab talks about #spatialtranscriptomic profiling of tissues infected with #SARS #COVID19 #AGBT21

Sharing our experience about spatial transcriptomic in thyroid lesions. @ThaisMaloberti #spatialtranscriptomic #thyroidtumors

Our summit in Paris is running successfully today; thanks to all our great speakers #spatialtranscriptomic

The first installation of @nanostringtech #cosmx in the UK 🇬🇧 explains all the big smiles! Looking forward to the incredible #singlecell #spatialtranscriptomic data that @LabSpatialNBJ team will produce @UofGCancerSci @UofGMVLS @UofGlasgow Thanks @The_MRC @CRUKresearch



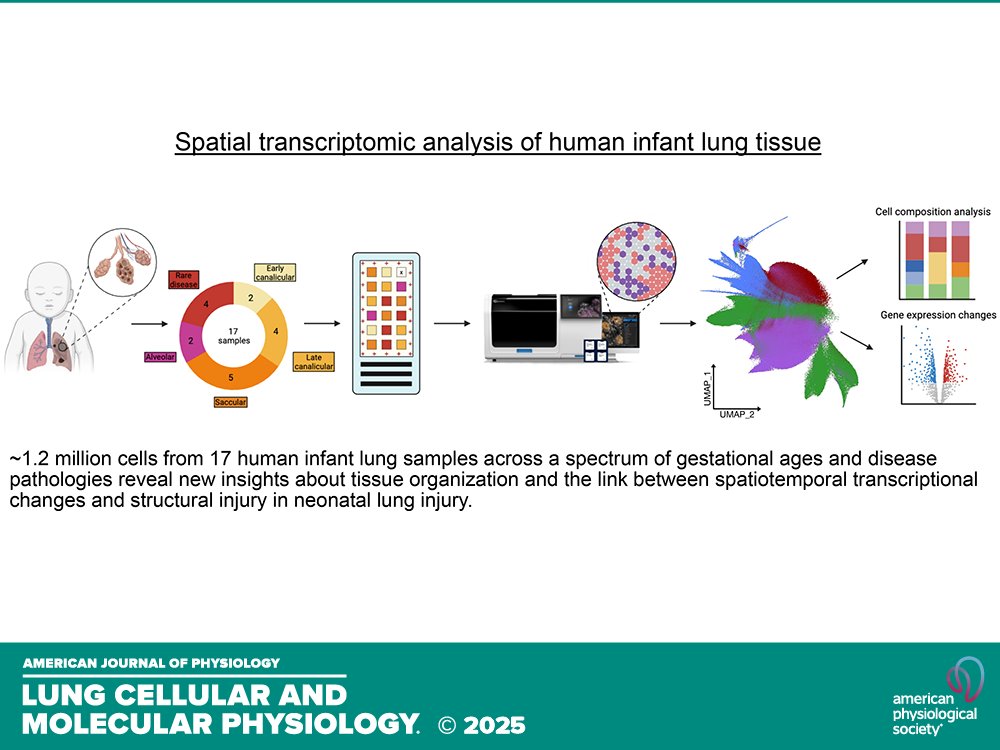
Short Report by S Mallapragada et al. (@AnnikaVannan @ASU @TGenResearch @VUMChealth @UW) A #spatialtranscriptomic atlas of acute #neonatal #lunginjury across development and disease severity ow.ly/nYfo50WMbH5
#spatialtranscriptomic alert 🚨 The MERSCOPE @vizgen_inc arrived at our single cell facility @Institut_Curie (Initiative Single Cell). Many Curie’s teams are ready to play with it, with us and Kyra Borgman from @CoulonLab.

Last week has been the most exciting in our Spatial Genomic Team -at @sangerinstitute , for beta-testing the N-01 @10xGenomics #CytAssist with #Visium FFPE Version2 I’m so grateful and honoured to have actually run our samples in the first model!! 🔬👩🏻🔬❤️#spatialtranscriptomic




Have you had the chance to explore what a #MERFISHmeasurement dataset from our #MERSCOPE in situ #SpatialTranscriptomic platform looks like? Take a break today and access the MERFISH Mouse Brain Receptor Map! hubs.ly/H0SWCFH0
Spatial transcriptomic is everywhere ! 🤩 @10xGenomics @LEGO_Group @institut_thorax #spatialtranscriptomic #science #biology #fun #kids

Nice talk from Tarine Standgaard on tumour heterogeneity in #NMIBC & use of #SpatialTranscriptomic (& bulk) analyses to distinguish BCG-responsive molecular subtype So gd to finally see #EAUlab up and running!! Thanks #EAU25 committee & #ESUR @Uroweb @LauraBukavinaMD @LDyrskjot



Friday afternoon fun going through my first @nanostringtech #GeoMx images. Trying to optimise morphology markers before embarking on the first #SpatialTranscriptomic experiment to be run @cardiffuni

🎇For the first time ever, we propose Infinite #SpatialTranscriptomic editing using #GenerativeAI. Our approach enables algorithmic gene expression-guided editing in a generated gigapixel mouse pup🐭. Code, preprint and generated 106496 x 53248 WSIs: github.com/CTPLab/IST-edi…

Advice needed: can you distinguish splicing variants by commercial #spatialtranscriptomic (eg visium)? #singlecell #omics
Short Report by S Mallapragada et al. (@AnnikaVannan @ASU @TGenResearch @VUMChealth @UW) A #spatialtranscriptomic atlas of acute #neonatal #lunginjury across development and disease severity ow.ly/nYfo50WMbH5

Post-doctoral researcher in oncology and computer science Paris Brain Institute See the full job description on jobRxiv: jobrxiv.org/job/paris-brai… #NeuroOncology #radiomic #spatialtranscriptomic #ScienceJobs jobrxiv.org/job/paris-brai…
Fascinating study from prof @ZeminZhangLab from Peking University fresh out in @Nature using both #scrna & #spatialtranscriptomic to decipher cross-tissue multicellular coordination and rewiring in cancer!
Nature research paper: Cross-tissue multicellular coordination and its rewiring in cancer go.nature.com/3ZEdXYZ
Excited to share my latest @JHepatology editorial on this impactful @Nature paper by my good friends @JoeYeong & Prof Liu Lianxin—highlighting Singapore–China collaboration & the power of #spatialtranscriptomic in predicting #HCC #recurrence. sciencedirect.com/science/articl…
Nice talk from Tarine Standgaard on tumour heterogeneity in #NMIBC & use of #SpatialTranscriptomic (& bulk) analyses to distinguish BCG-responsive molecular subtype So gd to finally see #EAUlab up and running!! Thanks #EAU25 committee & #ESUR @Uroweb @LauraBukavinaMD @LDyrskjot



STMiner, an algorithm specifically designed for complex #spatialtranscriptomic data can remove false-positive genes from the analysis results. We are deeply appreciative of @CellGenomics for affording us the platform to present this significant research outcome.

Caractéristiques spatiales et sensibilité aux traitements de 10 indications tumorales. #spatialtranscriptomic. The spatial landscape of cancer hallmarks reveals patterns of tumor ecological dynamics and drug sensitivity: Cell Reports cell.com/cell-reports/f…
The latest technical blog from scientists at the Earlham Institute shares some insights, top tips, and a workflow for applying #spatialtranscriptomic technologies to plant tissue. 🔎🌿🧬 ➡️ okt.to/ujacFs #PlantScience

Our CYGNUS project is developing a powerful new pipeline for the detailed study of diagnostic and archival tissue samples, enabling researchers to explore #SpatialTranscriptomic data 🔬 Discover more at buff.ly/4eg0bky Image by Jamieson Spatial Lab

🧬 Scientists at the Earlham Institute are developing advanced #singlecell and #spatialtranscriptomic approaches to explore gene expression in plant and animal systems, uncovering the mechanisms they use to adapt to challenges. #CellularGenomics okt.to/e73nFf
Neuronal heterogeneity in the medial septum and diagonal band of Broca: classes and continua biorxiv.org/content/10.110… #MERFISH #spatialtranscriptomic #singlecell #RNA #MERSCOPE @vizgen_inc
Discover imaging solutions at @MedDiscCat, transforming drug discovery and enhancing therapeutic effectiveness. 💊 From #MassSpectrometryImaging to #SpatialTranscriptomic capability, unlock the potential of integrated data to elevate development. Read👉 loom.ly/utgKgk4

Sharing our experience about spatial transcriptomic in thyroid lesions. @ThaisMaloberti #spatialtranscriptomic #thyroidtumors

Excited to share this fantastic opportunity for an intensive hands-on course in #spatialtranscriptomic with the @10xGenomics #Xenium @MRC_WIMM @UniofOxford. Perfect for those looking to expand their skills - don’t miss out! Unlock the secrets and register today 👇 #SingleCellOmic
Join us for a three-day hands-on course in Spatial Transcriptomics with the 10x Xenium at the @MRC_WIMM Advanced Single Cell Omics Facility (WASCOF) @UniofOxford. From Panel design & Tissue Preparation to Imaging & Data Analysis. Register your intetest: [email protected]


Super excited to be a part of this amazing team, and looking forward to discovering more with the cutting edge #spatialtranscriptomic for lung cancer patients.
Grateful for the support from the MRFF and incredible researchers supporting this Lung Cancer #MetaSpatial study #teamscience @chinwee10 @Dr_MarkAdams @bhuva_dd @lnly0311 @ProfGabBelz @CCRG_Research @Qld_SBC @WesleyResearch @WEHI_research @TRI_info @SAiGENCI @UQ_News
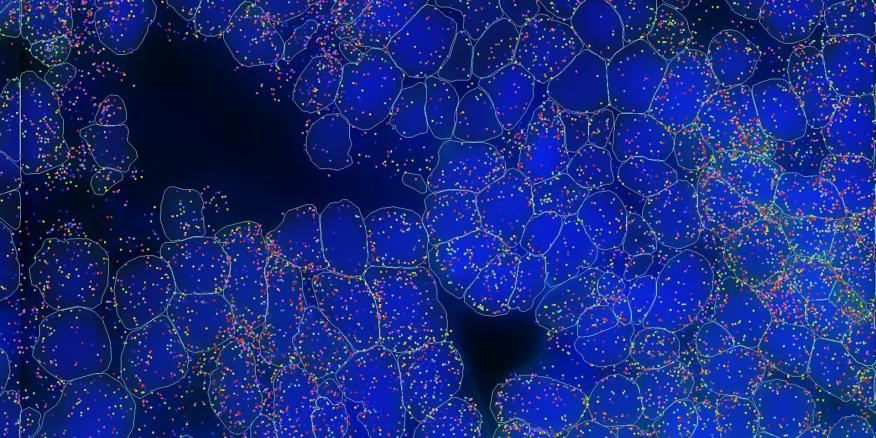
📷#MERSCOPE generates #MERFISH #SpatialTranscriptomic #DataImages that are not only exceptional in their #Accuracy & #DetectionEfficiency, but also breathtaking in their beauty.🎨 Check out the winners of our 2024 calendar contest: hubs.ly/Q02k1LM40 #VizgenViews #SciArt

🎇For the first time ever, we propose Infinite #SpatialTranscriptomic editing using #GenerativeAI. Our approach enables algorithmic gene expression-guided editing in a generated gigapixel mouse pup🐭. Code, preprint and generated 106496 x 53248 WSIs: github.com/CTPLab/IST-edi…

Join us in Seattle on 10/26 at the @10xGenomics User Group Meeting for presentations from leading scientists & networking! Visit our table for a demo of Partek Flow and see how to easily analyze your #singlecell and #spatialtranscriptomic data. partek.com/10x_10.26.23_S…



A🐭🧠 project spearheaded by @AllenInstitute created a #SpatialTranscriptomic #CellTypeAtlas of the whole #MouseBrain! Many key marker genes used in our new #MERSCOPE PanNeuro Cell Type Panel were validated in this project. Read the paper & learn more: hubs.ly/Q01KGWK90

Thrilled to receive the NIH/NIGMS R35 MIRA to support our #bioinformatics work studying subcellular and cellular #spatialtranscriptomic heterogeneity Special thanks to @rajivmccoy for guidance + support from mentors @jtleek, Joel Bader, Mike Miller @JHUBME 🥳🥂 #AcademicChatter

Check out 🔻🔻 #cholangiocarcinoma Paper from @chiarabraconi @ColmJJORourke gut.bmj.com/content/early/… Well done @LabSpatialNBJ for #spatialtranscriptomic component @Gut_BMJ @grisurgery @nanostringtech @UofGMVLS @UofGCancerSci


@mason_lab talks about #spatialtranscriptomic profiling of tissues infected with #SARS #COVID19 #AGBT21

In December 2020, Dr. Zhuang's lab at @Harvard posted an article to @biorxivpreprint detailing how they used #MERFISH to generate a subcellular #SpatialTranscriptomic atlas of neurons. Read & learn about the patterns of #RNA in neurons. hubs.ly/H0-cgx90 #ThrowbackThursday

Short Report by S Mallapragada et al. (@AnnikaVannan @ASU @TGenResearch @VUMChealth @UW) A #spatialtranscriptomic atlas of acute #neonatal #lunginjury across development and disease severity ow.ly/nYfo50WMbH5

Welcome to Tu Duyen Nguyen, @AllianceProg intern from @Polytechnique. Tu Duyen is joining the IICD to focus on novel algorithms to understand the spatial interaction between cells from #spatialtranscriptomic data in the @elhamazizi lab.

Starting today....@nanostringtech Nanostring GEOMx fully operational and training beginning! @UofGMVLS @pvanhouts Mathias Holpert @UofGCancerSci @CRUKGlasgow looking forward to exciting #spatialtranscriptomic projects @CRUK_BI @grisurgery @hollyleslie_ @EdinCRC @precisionpanc




1/4 #SpatialTranscriptomic methods allow to analyse the gene expression of tissues without losing its spatial heterogeneity. In my #PhD in the Meier lab @PioneerCampus I apply and advance such methods to address different biological questions. #YoungScientistsHMGU

📷#MERSCOPE generates #MERFISH #SpatialTranscriptomic #DataImages that are not only exceptional in their #Accuracy & #DetectionEfficiency, but also breathtaking in their beauty.🎨 Check out the winners of our 2024 calendar contest: hubs.ly/Q02k1LM40 #VizgenViews #SciArt

The first installation of @nanostringtech #cosmx in the UK 🇬🇧 explains all the big smiles! Looking forward to the incredible #singlecell #spatialtranscriptomic data that @LabSpatialNBJ team will produce @UofGCancerSci @UofGMVLS @UofGlasgow Thanks @The_MRC @CRUKresearch



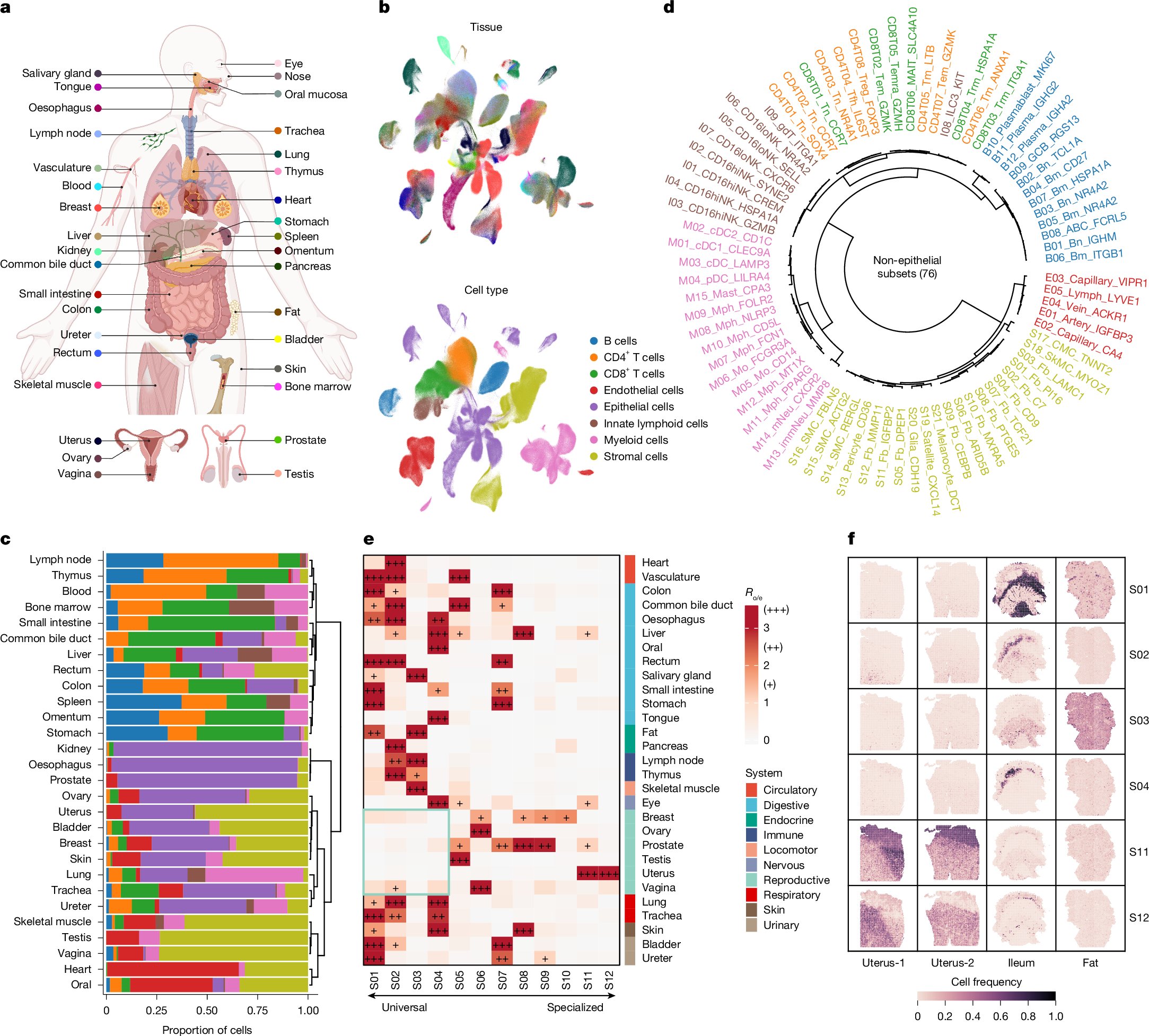
The first in-depth reference map of the uterus to collect both single cell and #spatialtranscriptomic data reveals new insights into the cellular make up of #endometrialcancers and adds to the Human Cell Atlas. Learn more: ow.ly/f5Xz50H35bQ #uterinecancer
Now open! Research Award for #SpatialBiology across the #WholeTranscriptome. Two awards available: Human & Mouse. #SpatialTranscriptomic. In partnership with @EACRnews and @illumina. Apply today! ow.ly/Bfig50EwdaV

Nice talk from Tarine Standgaard on tumour heterogeneity in #NMIBC & use of #SpatialTranscriptomic (& bulk) analyses to distinguish BCG-responsive molecular subtype So gd to finally see #EAUlab up and running!! Thanks #EAU25 committee & #ESUR @Uroweb @LauraBukavinaMD @LDyrskjot



Spatial transcriptomic is everywhere ! 🤩 @10xGenomics @LEGO_Group @institut_thorax #spatialtranscriptomic #science #biology #fun #kids

Single Cell RNA-Seq and Spatial Data Merge Using Cell2location A team has developed a tool, сell2location, that can resolve fine-grained cell types in #spatialtranscriptomic data and create comprehensive cellular maps of diverse tissues. Learn more: ow.ly/9Zya50HuEaq

Last week has been the most exciting in our Spatial Genomic Team -at @sangerinstitute , for beta-testing the N-01 @10xGenomics #CytAssist with #Visium FFPE Version2 I’m so grateful and honoured to have actually run our samples in the first model!! 🔬👩🏻🔬❤️#spatialtranscriptomic




Rao's team at the @QIMRBerghofer used GeoMx DSP to study #spatialtranscriptomic signatures and viral load in lungs from #SARS-CoV-2 infected hamsters and revealed that nuclear ACE2 may be a therapeutic target independent of the variant. 👉go.nature.com/42ZfAiR

@OliverStegle presenting on identification of spatially variable genes #spatialtranscriptomic #cytodata2019

#spatialtranscriptomic alert 🚨 The MERSCOPE @vizgen_inc arrived at our single cell facility @Institut_Curie (Initiative Single Cell). Many Curie’s teams are ready to play with it, with us and Kyra Borgman from @CoulonLab.

Something went wrong.
Something went wrong.
United States Trends
- 1. Gibbs 7,167 posts
- 2. #WWERaw 10.9K posts
- 3. Lions 53.1K posts
- 4. $BIGMI 1,607 posts
- 5. Bucs 11.4K posts
- 6. #OnePride 3,320 posts
- 7. Ben Solo 7,685 posts
- 8. East Wing 36.1K posts
- 9. FanDuel 21.4K posts
- 10. Goff 5,211 posts
- 11. Mike Evans 2,127 posts
- 12. Jack Campbell N/A
- 13. #MIRXtakeoff N/A
- 14. Bron 16.2K posts
- 15. Alim 7,171 posts
- 16. Brad Allen 1,293 posts
- 17. Cobie 13.6K posts
- 18. Tyler Lockett 1,591 posts
- 19. Greenlaw 2,966 posts
- 20. #TBvsDET 1,404 posts