#toolsbenchmarking search results
Benchmarking computational methods for B-cell receptor reconstruction from single-cell RNA-seq data. #scRNAseq #BcellReceptor #ToolsBenchmarking #NARgenomicsAndBioinformatics academic.oup.com/nargab/article…
academic.oup.com
Benchmarking computational methods for B-cell receptor reconstruction from single-cell RNA-seq data
Abstract. Multiple methods have recently been developed to reconstruct full-length B-cell receptors (BCRs) from single-cell RNA sequencing (scRNA-seq) data
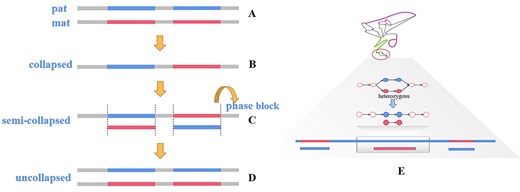
Complex genome assembly based on long-read sequencing' #GenomeAssembly #LongReads #ToolsBenchmarking @BriefingBioinfo #Bioinformatics academic.oup.com/bib/advance-ar…
Comparison of scRNA-seq data analysis method combinations. #scRNAseq #DataAnalysis #ToolsBenchmarking #Bioinformatics #BriefingsInFunctionalGenomics academic.oup.com/bfg/advance-ar…
academic.oup.com
Comparison of scRNA-seq data analysis method combinations
Abstract. Single-cell ribonucleic acid (RNA)-sequencing (scRNA-seq) data analysis refers to the use of appropriate methods to analyze the dataset generated
A benchmark of computational pipelines for single-cell histone modification data. #SingleCell #HistoneModifications #ToolsBenchmarking @GenomeBiology genomebiology.biomedcentral.com/articles/10.11…
Benchmarking and integration of methods for deconvoluting spatial transcriptomic data. #SpatialTransciprtomics #ToolsBenchmarking #Bioinformatics academic.oup.com/bioinformatics…
A comprehensive benchmarking of WGS-based deletion structural variant callers. #StructuralVariants #SV #ToolsBenchmarking #GoldStandardSVdatasets #Bioinformatics @BriefingBioinfo academic.oup.com/bib/advance-ar…
academic.oup.com
A comprehensive benchmarking of WGS-based deletion structural variant callers
Abstract. Advances in whole-genome sequencing (WGS) promise to enable the accurate and comprehensive structural variant (SV) discovery. Dissecting SVs from
Detection of oncogenic and clinically actionable mutations in cancer genomes critically depends on variant calling tools. #VariantCalling #ToolsBenchmarking #VariantAnalysis @Alfons_Valencia @eportacangen academic.oup.com/bioinformatics… #Bioinformatics
Performance evaluation of computational methods for splice-disrupting variants and improving the performance using the machine learning-based framework. #ExomeSplicingVariants #ToolsBenchmarking #MachineLearning #Bioinformatics @BriefingBioinfo academic.oup.com/bib/advance-ar…
A systematic comparison of human mitochondrial genome assembly tools. #HumanMitochondrialGenome #GenomeAssembly #ToolsBenchmarking @BMCBioinfo bmcbioinformatics.biomedcentral.com/articles/10.11…
Accuracy and completeness of long read metagenomic assemblies. #MetagenomicsAssembly #ToolsBenchmarking #Bioinformatics @biorxiv_bioinfo biorxiv.org/content/10.110…
Comprehensive assessment of differential ChIP-seq tools guides optimal algorithm selection. #ChiPseq #ToolsBenchmarking #ReferenceDatasets #Bioinformatics genomebiology.biomedcentral.com/articles/10.11… @GenomeBiology
Evaluation of different computational methods for DNA methylation-based biological age. #ToolsBenchmarking #DNAmethylation #BiologicalAge #Bioinformatics @BriefingBioinfo academic.oup.com/bib/advance-ar…
Systematic evaluation of cell-type deconvolution pipelines for sequencing-based bulk DNA methylomes. #ToolsBenchmarking #DNAMethylation #Sequencing #Bioinformatics @BriefingBioinfo academic.oup.com/bib/advance-ar… @
Benchmarking of bioinformatics tools for the hybrid de novo assembly of human whole-genome sequencing data. #WGS #GenomeAssembly #ToolsBenchmarking @LabCflores @biorxivpreprint biorxiv.org/content/10.110…
Graph construction method impacts variation representation and analyses in a bovine super-pangenome. #BovinePangenomes #ToolsBenchmarking @biorxivpreprint biorxiv.org/content/10.110…
Systematic benchmarking of statistical methods to assess differential expression of circular RNAs. #DifferentialExpression #CircularRNA #ToolsBenchmarking #Bioinformatics @BriefingBioinfo academic.oup.com/bib/advance-ar…
academic.oup.com
Systematic benchmarking of statistical methods to assess differential expression of circular RNAs
Abstract. Circular RNAs (circRNAs) are covalently closed transcripts involved in critical regulatory axes, cancer pathways and disease mechanisms. CircRNA
Comprehensive evaluation of methods for differential expression analysis of metatranscriptomics data. #MetaTranscriptomics #ToolsBenchmarking @biorxivpreprint biorxiv.org/content/10.110…
Benchmarking Automated Cell Type Annotation Tools for Single-cell ATAC-seq Data. #scATACseq #SingleCellAnnotation #ToolsBenchmarking @biorxivpreprint biorxiv.org/content/10.110…
A benchmarking of human Y-chromosomal haplogroup classifiers from whole-genome and whole-exome sequence data. #YchrClassifiers # WGS #WES #ToolsBenchmarking @biorxivpreprint biorxiv.org/content/10.110…
Benchmarking of bioinformatics tools for the hybrid de novo assembly of human whole-genome sequencing data. #WGS #GenomeAssembly #ToolsBenchmarking @LabCflores @biorxivpreprint biorxiv.org/content/10.110…
A systematic comparison of human mitochondrial genome assembly tools. #HumanMitochondrialGenome #GenomeAssembly #ToolsBenchmarking @BMCBioinfo bmcbioinformatics.biomedcentral.com/articles/10.11…
A benchmark of computational pipelines for single-cell histone modification data. #SingleCell #HistoneModifications #ToolsBenchmarking @GenomeBiology genomebiology.biomedcentral.com/articles/10.11…
Systematic benchmarking of statistical methods to assess differential expression of circular RNAs. #DifferentialExpression #CircularRNA #ToolsBenchmarking #Bioinformatics @BriefingBioinfo academic.oup.com/bib/advance-ar…
academic.oup.com
Systematic benchmarking of statistical methods to assess differential expression of circular RNAs
Abstract. Circular RNAs (circRNAs) are covalently closed transcripts involved in critical regulatory axes, cancer pathways and disease mechanisms. CircRNA
Comprehensive evaluation of methods for differential expression analysis of metatranscriptomics data. #MetaTranscriptomics #ToolsBenchmarking @biorxivpreprint biorxiv.org/content/10.110…
Benchmarking and integration of methods for deconvoluting spatial transcriptomic data. #SpatialTransciprtomics #ToolsBenchmarking #Bioinformatics academic.oup.com/bioinformatics…
Accuracy and completeness of long read metagenomic assemblies. #MetagenomicsAssembly #ToolsBenchmarking #Bioinformatics @biorxiv_bioinfo biorxiv.org/content/10.110…
Graph construction method impacts variation representation and analyses in a bovine super-pangenome. #BovinePangenomes #ToolsBenchmarking @biorxivpreprint biorxiv.org/content/10.110…
A benchmarking of human Y-chromosomal haplogroup classifiers from whole-genome and whole-exome sequence data. #YchrClassifiers # WGS #WES #ToolsBenchmarking @biorxivpreprint biorxiv.org/content/10.110…
Benchmarking Automated Cell Type Annotation Tools for Single-cell ATAC-seq Data. #scATACseq #SingleCellAnnotation #ToolsBenchmarking @biorxivpreprint biorxiv.org/content/10.110…
Comparison of scRNA-seq data analysis method combinations. #scRNAseq #DataAnalysis #ToolsBenchmarking #Bioinformatics #BriefingsInFunctionalGenomics academic.oup.com/bfg/advance-ar…
academic.oup.com
Comparison of scRNA-seq data analysis method combinations
Abstract. Single-cell ribonucleic acid (RNA)-sequencing (scRNA-seq) data analysis refers to the use of appropriate methods to analyze the dataset generated
Performance evaluation of computational methods for splice-disrupting variants and improving the performance using the machine learning-based framework. #ExomeSplicingVariants #ToolsBenchmarking #MachineLearning #Bioinformatics @BriefingBioinfo academic.oup.com/bib/advance-ar…
Complex genome assembly based on long-read sequencing' #GenomeAssembly #LongReads #ToolsBenchmarking @BriefingBioinfo #Bioinformatics academic.oup.com/bib/advance-ar…
Something went wrong.
Something went wrong.
United States Trends
- 1. Happy Thanksgiving Eve 3,010 posts
- 2. Good Wednesday 25.5K posts
- 3. #wednesdaymotivation 4,053 posts
- 4. Colorado State 2,996 posts
- 5. Nuns 7,012 posts
- 6. #Wednesdayvibe 1,961 posts
- 7. Mora 21.3K posts
- 8. Happy Hump 6,243 posts
- 9. Hump Day 9,384 posts
- 10. Luka 68.2K posts
- 11. Lakers 53.4K posts
- 12. Clippers 19.7K posts
- 13. Collar 47.3K posts
- 14. Karoline Leavitt 25.4K posts
- 15. Tina Turner 4,775 posts
- 16. The God 413K posts
- 17. #StrangerThings5 16.1K posts
- 18. Periods 13.9K posts
- 19. Witkoff 175K posts
- 20. Joy Reid 4,518 posts