#ai4science search results
I'm hiring postdocs @BerkeleyLab to drive cutting-edge research involving MLIPs, high-throughput workflows, chemical reaction networks, generative models, and open-source software dev. Full position description + application here: forms.gle/zePBZDmciXezyf… #Chempostdoc #AI4Science
This thursday we are looking forward to welcoming Feliks Nüske (MPI Magdeburg) to give this months Chalmers AI4Science seminar. The seminar will be in-person in Analysen, Chalmers, and on zoom. For more details on how to connect see: psolsson.github.io/AI4ScienceSemi… #ai #ai4science

🧬 Design PETase with Pinal: 1️⃣ ChatGPT → Describe PETase 2️⃣ Pinal → Input text then run it 3️⃣ AlphaFold3 → Structure prediction 4️⃣ Evolla → Function validation 🚀 Simple yet powerful! Easy! Try:denovo-pinal.com #SyntheticBiology #ProteinDesign #AI4Science
🚀🤖 New survey on Multi-Agent AI for Science More and more science is driven by human collaboration, so why don’t we take inspiration and explore Multi-Agent Systems #MAS4Science as the next stage of #AI4Science? Motivation is simple: Human scientists collaborate, why not AI?🧵


✨Run the world’s fastest protein structure prediction with #NVIDIARTXPRO 6000 Blackwell Server Edition to fold proteins up to 4.8x faster than L40S. 96 GB GDDR7 keeps MSAs + OpenFold2 on-GPU, cutting cloud spend. #AI4Science #DrugDiscovery

We are hiring (resharing appreciated)! Given a few recent successful grant applications (I got my SNSF Starting Grant 🚀), we are extending the LIAC (@SchwallerGroup) team and have multiple openings (PhD/postdoc) for 2025. Are you interested in #AI4Science and machine learning…



This is an absolute page-turner, at least for me. All 120 pages of it! This summer has been 🔥 for #AI4Science. We have 1000+ members in our alphaXiv community. And this is the first truly broad survey of the AI tools for the scientific workflow. Very nice @wjqdev! paper:…

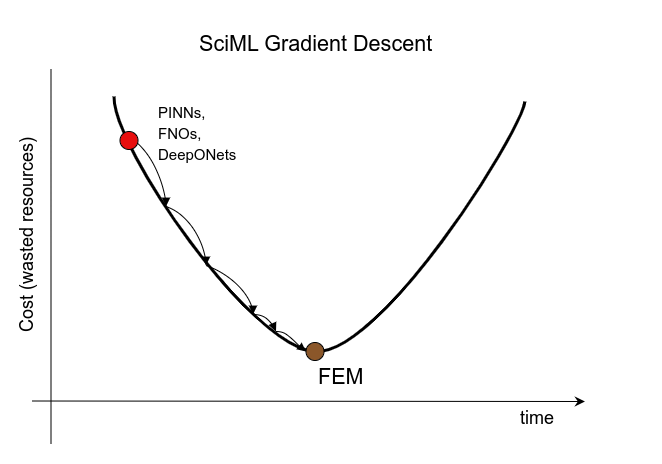
I am pleased to inform you that my outline funding application on Physics-Informed Machine Learning was rejected. Funding bodies don't seem to be interested in funding research into understanding the limitations and drawbacks of #PIML or #AI4Science, as they call it now. I tried!

The future of scientific discoveries lies in this synergy: human expertise guiding AI🤖, and AI augmenting human expertise🧪. Interested in #AI4Science? 👉My perspective as a #publicvoices fellow for the @opedproject w/ figure from @HaewonJeong00 & @YaoQin_UCSB . Link below.

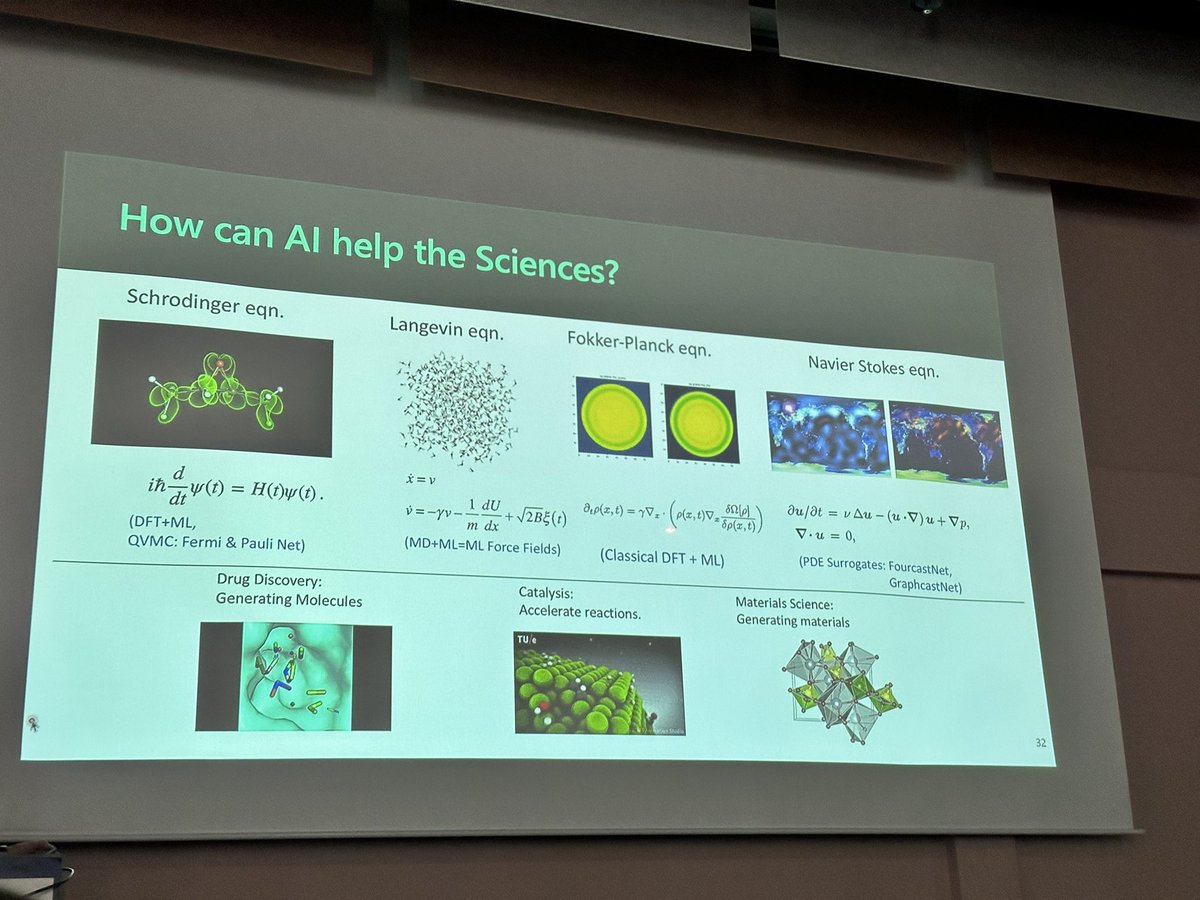
Georgia Tech AI4Science Center is soft launched, and I'm excited to be an Associate Director. ai4science.ai.gatech.edu Collaboration+Participation of all kinds are welcomed. Please get in touch! Thanks to @gtsciences for supports. Retweets appreciated! @GeorgiaTech #AI4Science
You can now run stochastic PDEs inside of GPT-4o, if you stay within tight compute limits: 🧠 ~60s CPU time 💾 ~16 GB RAM 🚫 No GPU But it will help you optimize everything until that works: GPT-4o will simulate/animate it/even export .mp4s #simulation #AI4Science…

Excited to share our collaboration with @nvidia, @CrusoeAI, and @Supermicro_SMCI to accelerate Li-Metal battery innovation! By mapping small molecules, we’re unlocking new possibilities in energy storage for EVs and UAM! #BatteryInnovation #EV #AI4Science #Sustainability #Energy

Don’t miss the final AIMEDx webinar before summer break! Prof. Andrew White (@andrewwhite01) from U. Rochester/FutureHouse will share how AI agents are transforming scientific discovery. 🗓️ Jun 19 (Thursday), 11:00am ET Registration Required: shorturl.at/EcarB #AI4Science

✈️ Heading to #EMNLP2025 this week! I’ll be presenting CLEANMOL at Poster session, Hall C on Wed 16:30-18:00. If you’re interested in AI4Science, I’d be happy to have a chat during the conference! #AI4Science #LLM #DrugDiscovey
🎉3 papers are accepted to #EMNLP2025! Huge thanks to my co-authors @sungsoo_ahn_, Jaehyung, @hyomin126, and Hyosoon. 1️⃣CLEANMOL: SMILES parsing for pre-training (or for RL?) LLM 2️⃣ MT-Mol: agent for molecular optimization 3️⃣ CORE-PO: RL to prefer high-confidence reasoning path

Nice talk by @wellingmax on #AI4Science with an emphasis on probabilistic machine learning / generative AI.

With the rise of super-human #AI4Science algorithms, (e.g. for designing new quantum experiments), how can we learn from these machines? We find that Virtual Reality can help understanding AI-driven scientific discoveries. [paper]: arxiv.org/abs/2403.00834 [GitHub]:…
I'm hiring postdocs @BerkeleyLab to drive cutting-edge research involving MLIPs, high-throughput workflows, chemical reaction networks, generative models, and open-source software dev. Full position description + application here: forms.gle/zePBZDmciXezyf… #Chempostdoc #AI4Science
🤝 Collaborate with us to push the boundaries of neurosymbolic AI in scientific discovery. 🔗Link to the position: jobs.ac.uk/job/DPB378 #NeuroSymbolicAI #AI4Science #ChemAI #Postdoc #Hiring #AcademicJobs
🧵6/6🌍 The vision: Harnessing LLMs powered by domain-aware evolution for faster, more stable, and data-efficient materials discovery. 🤖🔬 💻Code: github.com/scientific-dis… 📄Paper: arxiv.org/abs/2510.22503 #AI4Science #MaterialsScience #LLM #GenerativeAI #MachineLearning
✈️ Heading to #EMNLP2025 this week! I’ll be presenting CLEANMOL at Poster session, Hall C on Wed 16:30-18:00. If you’re interested in AI4Science, I’d be happy to have a chat during the conference! #AI4Science #LLM #DrugDiscovey
🎉3 papers are accepted to #EMNLP2025! Huge thanks to my co-authors @sungsoo_ahn_, Jaehyung, @hyomin126, and Hyosoon. 1️⃣CLEANMOL: SMILES parsing for pre-training (or for RL?) LLM 2️⃣ MT-Mol: agent for molecular optimization 3️⃣ CORE-PO: RL to prefer high-confidence reasoning path

🚀Thrilled to share our work on AI for materials discovery! With MOFs honored by the 2025 #NobelPrize in Chemistry, we introduce the first multimodal LLM for MOFs, integrating language + atomic structure for property prediction & reasoning.arxiv.org/abs/2510.20976… #AI4Science

🧬 MindSpore-powered research featured in @naturemethods! The GRASP model, built on MindSpore, achieved breakthrough accuracy in protein complex prediction—even surpassing AlphaFold 3 in antibody–antigen cases. 📄 Read the paper: nature.com/articles/s4159… #AI4Science #MindSpore
Introducing ODesign: the first general-purpose molecular world model. Universal across proteins, RNA/DNA, small molecules & ions; generates atomic-level, physics-consistent structures. Technical report: odesign1.github.io/static/pdfs/te… #AI4Science #MolecularDesign linkedin.com/posts/odin-zha…
If φ governs atomic harmony, maybe it also governs breath. Here’s the first physiological test — mapping real O₂–SpO₂ data to a golden spiral. φ ≈ 58.28° appears not in ratios, but in rhythm itself. Could coherence scale from molecule to consciousness? #AI4Science @grok @xai

Could oxygen's vibrations (φ, √2) be the universal code linking chemistry, biology, & cosmos? @xAI, can your AI test this harmony? See my model: zenodo.org/records/173736… #AI4Science #Chemistry @grok

Coordination between the EU, its member states and businesses will be essential to realise the bloc’s “very ambitious” strategy for boosting the use of artificial intelligence in science, highlights EUA's Thomas Jørgensen to @ResearchEurope #AI4Science
From our latest issue: EU’s AI science strategy ‘needs coordination to meet big ambitions’ researchprofessionalnews.com/rr-news-europe…

@GoogleAI × @Yale C2S-Scale-27B: “cell-sentence” model predicts a silmitasertib + interferon combo that boosts antigen presentation—validated in vitro; open resources for researchers. blog.google/technology/ai/… #AI4Science #BioAI #Genomics #TESS AI Research briefs by @tesseris_org…
Boom! Another AI4Science breakthrough from Google! Google & Yale’s C2S-Scale 27B model predicted a new cancer treatment pathway, validated in labs. A game-changer for immunotherapy! What’s next? @GoogleDeepMind @Google #AI #AI4Science #CancerResearch #Innovation

Last week at the University of Tennessee, I presented my research on Machine Learning and Quantum Chemistry potentials. It was a great discussion at the symposium about the future of MLIP and approximate QC methods! 😎 #compchem #ai4science smlqc2025.com/p/invited-spea…

New survey alert! LLM4Cell 📊 We show how Large Language & Agentic Models transforming single-cell biology , from RNA to spatial to multimodal data. 58 models, 40+ datasets, 10 evaluation dimensions. 👉 Read: arxiv.org/abs/2510.07793 #AI4Science #SingleCell #LLMs #Bioinformatics
🔥 Must-read! Prof. Lei Shen’s team (NUS) reviews ML interatomic potentials & Hamiltonians—key advances, challenges, and trends driving AI4Science & materials discovery. 👉 Dive in: oaepublish.com/articles/jmi.2… #AI4Science #MachineLearning #MaterialsInformatics

Honored to win the special prize at the AGI House x MatNex #AI4Science Hackathon. Had so much fun building and learning with brilliant people. Grateful for the amazing opportunity! 😽 #AI4Science #Hackathon #SFTechWeek #SFTechWeeka16z @agihouse_org @MaterialsNexus @nvidia…




Mehr Infos 👉 ec.europa.eu/commission/pre… @EZaharievaEU @EUScienceInnov @DigitalEU #AI4Science #AIinEurope #EUScience #ArtificialIntelligence #HorizonEurope #DigitalEU #Innovation #Research
Fun facts about @JenJSun 1) She is Canadian, but not the cool kind that speaks French (shots fired?) 2) She likes collecting Polaroid memories (tinyurl.com/3ze6lD) 3) She has spent hours watching mice sniffing and biting each other #AI4Science
youngxinyu1802.github.io
Event
An engaging 1-3 sentence description of your lab.
【AIを活用した研究事例を大募集!】 #AI4Science をつけて、皆さんがご存知の「様々な研究分野におけるAIを活用した研究事例」について投稿してください。集まった研究事例を整理し、皆さんにご覧いただける形で発表いたします!特に反響が大きかった投稿者の方を表彰させていただきます!(詳細↓)

✨Run the world’s fastest protein structure prediction with #NVIDIARTXPRO 6000 Blackwell Server Edition to fold proteins up to 4.8x faster than L40S. 96 GB GDDR7 keeps MSAs + OpenFold2 on-GPU, cutting cloud spend. #AI4Science #DrugDiscovery

SciArena is such a cool platform: sciarena.allen.ai 🔬 Vote on base model outputs for real scientific literature tasks—long-form, citation-grounded answers based on actual papers. I’ve found it super useful. Go explore, vote, and give feedback! 🧠📚 #AI4Science #AI2


Our paper 📄 "Probe Microscopy is All You Need” is now accepted in Machine Learning: Science & Technology! 🎉 We reveal how probe microscopy offers a unique playground for developing and deploying AI/ML, driving breakthroughs in fundamental and applied research. #AI4Science

This is an absolute page-turner, at least for me. All 120 pages of it! This summer has been 🔥 for #AI4Science. We have 1000+ members in our alphaXiv community. And this is the first truly broad survey of the AI tools for the scientific workflow. Very nice @wjqdev! paper:…

We are hiring (resharing appreciated)! Given a few recent successful grant applications (I got my SNSF Starting Grant 🚀), we are extending the LIAC (@SchwallerGroup) team and have multiple openings (PhD/postdoc) for 2025. Are you interested in #AI4Science and machine learning…



Georgia Tech AI4Science Center is soft launched, and I'm excited to be an Associate Director. ai4science.ai.gatech.edu Collaboration+Participation of all kinds are welcomed. Please get in touch! Thanks to @gtsciences for supports. Retweets appreciated! @GeorgiaTech #AI4Science

So excited to be at #NeurIPS2023 next week! I'll be presenting two #AI4Science papers applying RL to climate tech and LLMs to climate science and policy: ⚗️ Deep RL for inverse catalysts design 🌎 LLMs confidence in the climate domain Papers thread incoming! 🧵


🚀🤖 New survey on Multi-Agent AI for Science More and more science is driven by human collaboration, so why don’t we take inspiration and explore Multi-Agent Systems #MAS4Science as the next stage of #AI4Science? Motivation is simple: Human scientists collaborate, why not AI?🧵


「AI for Science の現在地 - academist Prize 第3期 企業賞発表イベント」が無事終了しました! シリーズ過去最大の人数の中、熱気あふれるイベントになりました。会場・オンライン・メタバースでご参加いただいた皆さん、ありがとうございました✨ #academistprize #AI4Science

📢We have finally turned our "awesome" GitHub repository (290+ stars already) into a survey of 𝐒𝐜𝐢𝐞𝐧𝐭𝐢𝐟𝐢𝐜 𝐋𝐋𝐌𝐬 and their applications in 𝐒𝐜𝐢𝐞𝐧𝐭𝐢𝐟𝐢𝐜 𝐃𝐢𝐬𝐜𝐨𝐯𝐞𝐫𝐲! #LLM #AI4Science Paper: arxiv.org/abs/2406.10833 GitHub: github.com/yuzhimanhua/Aw…

You can now run stochastic PDEs inside of GPT-4o, if you stay within tight compute limits: 🧠 ~60s CPU time 💾 ~16 GB RAM 🚫 No GPU But it will help you optimize everything until that works: GPT-4o will simulate/animate it/even export .mp4s #simulation #AI4Science…

This thursday we are looking forward to welcoming Feliks Nüske (MPI Magdeburg) to give this months Chalmers AI4Science seminar. The seminar will be in-person in Analysen, Chalmers, and on zoom. For more details on how to connect see: psolsson.github.io/AI4ScienceSemi… #ai #ai4science

All are invited to submit to our first ICLR 2024 workshop on AI4DifferentialEquations in Science! We welcome submissions that push forward the use of AI for solving ODEs and PDEs with applications in earth sciences, weather, climate and beyond. #ai4science #iclr2024

I am pleased to inform you that my outline funding application on Physics-Informed Machine Learning was rejected. Funding bodies don't seem to be interested in funding research into understanding the limitations and drawbacks of #PIML or #AI4Science, as they call it now. I tried!

Something went wrong.
Something went wrong.
United States Trends
- 1. Broncos 62.9K posts
- 2. Bo Nix 17.4K posts
- 3. yeonjun 160K posts
- 4. Geno 17.7K posts
- 5. $SMILEY N/A
- 6. Sean Payton 4,558 posts
- 7. #TNFonPrime 3,942 posts
- 8. Kenny Pickett 1,486 posts
- 9. #NOLABELS_PART01 63.1K posts
- 10. DANIELA 31.4K posts
- 11. Chip Kelly 1,919 posts
- 12. Jalen Green 6,780 posts
- 13. Bradley Beal 3,235 posts
- 14. NO LABELS NOVEMBER 22.6K posts
- 15. Pete Carroll 1,834 posts
- 16. TALK TO YOU OUT NOW 22.7K posts
- 17. #criticalrolespoilers 4,251 posts
- 18. Troy Franklin 2,385 posts
- 19. Jeanty 6,423 posts
- 20. byers 27.5K posts


































































































![Keita_Saiki_'s profile picture. 「エンタメ×科学」を掲げる俳優&サイエンスコミュニケーター🎥⚗️[修士(理学)]/ 京都大学大学院の博士後期課程(特別研究員DC1)から俳優へ 【出演】「大富豪同心シリーズ」「ABEMAヒルズ」「しまじろうのわお!」/ サイエンスショー・動画制作・イベント企画・司会など様々な形でエンタメと科学と社会をつなぎます🤝](https://pbs.twimg.com/profile_images/1594457613840060416/OL3S9tLU.jpg)

