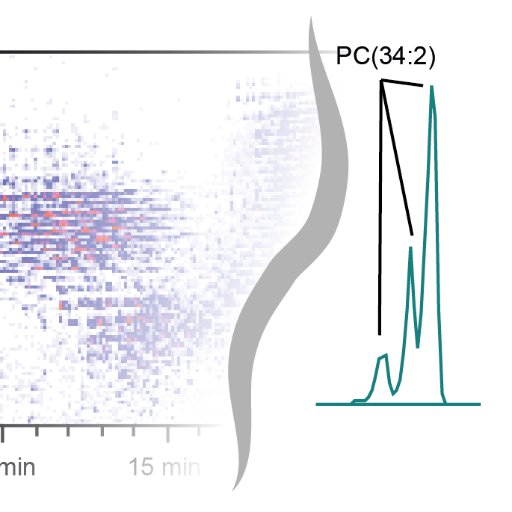
#compms search results
Today's the last day of @iscb's virtual flagship conference - it's been a wonderful experience exploring the computational work everyone has shared at #ISMB2020! I'll be "by my poster" in the #CompMS COSI this morning 🤓 #proteomics #computational #MassSpec #MachineLearning

Right now in the CompMS cosi at ISMB is "software solutions for multi-omic research" by Evgenia Shiskova from university of Wisconsin #ISMB18 #CompMS
It’s time! @iscb‘s ISMB2020 is starting tomorrow! I’ve got a poster and brief presentation (#552) in the #computational mass spec track 🤓 as do many other members of the @mlaval6 lab 🔥 #compMS #ISMB2020

Our #compMS workshop at @OISTedu was very well received. Thanks to @rschmid1789 @corinnabrungs @roman_bushuiev @damiani_tito @LF_Nothias @boeckerlab!


TagGraph: Match de novo peptides to sequence databases. Fast! ✨String-based ✨No precursor assumptions ✨No digestion assumptions ✨No PTM assumptions ✨FDR estimation #ISMB18 #CompMS #massspec #proteomics #PTMs #denovosequencing #databasesearch



We're kicking off the #CompMS session at the #ismbeccb2021 conference with a keynote presentation by @thalexandrov on how computational MS enables spatial and single-cell metabolomics. Ready for two days of #CompMS goodness!

Really cool session of introduction to matchms Python package and Jupyter notebooks itself. Very easy to follow the lesson with this well-written google colab notebook. This is a really powerful package for playing with msms data! @mzmine_project #compMS @vdHooft_CompMet

Last keynote of #CompMS COSI at #ISMB2018 by Josh Elias on some cool de novo search stuff (TagGraph)...

We are excited to announce our summer 2022 fellowship!! Great opportunity for grad students with #MassSpec background to receive training in #CompMS. The application deadline is May 6. #AcademicTwitter #opensource #proteomics #Metabolomics #Lipidomics #TeamMassSpec

Hannes Röst opening the #CompMS COSI track at #ISMBECCB 2019 in Basel with a talk on DIA data analysis

This workshop at @IOCBPrague is going to be a great opportunity to learn about the latest developments in @mzmine_project and other #compMS tools!

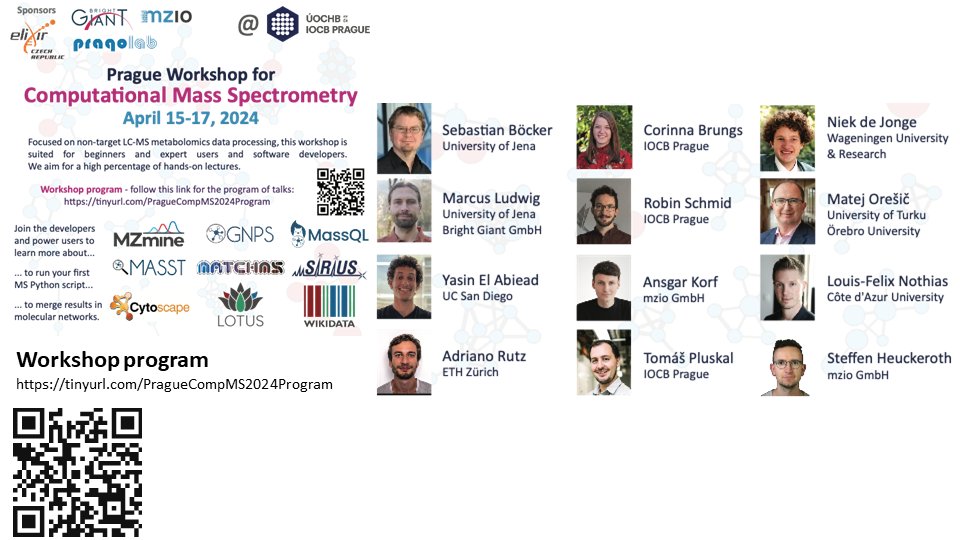
Register now! Prague Computational Mass Spectrometry Workshop, April 15-17, 2024, Prague, Czechia. Learn from the developers and power users of @mzmine_project, SIRIUS, #matchms, @TheLOTUSInitia1, @GNPS_UCSD and discuss how we can move the field forward... tinyurl.com/PragueCompMS20…
Great talk by Johanna Galvis on an open source tool for differential analysis of isotope-labeled metabolomics data #ISMBECCB2023 #compMS

Final day of #ISMBECCB2023 came with the wonderful invited keynote from Bernhard @RenardLab on his journey with spectra and their current and outlook! #CompMS




Ready for the Prague Computational Mass Spectrometry Workshop at the @IOCBPrague. Thanks to the organizers @mzmine_project @tomas_pluskal @rschmid1789 and others #compMS




@sneumannoffice presenting the R #metabolomics world at the #CompMS #computational #metabolomics COSI #ismb2020 track!


Now it is my turn at the #CompMS #computational #metabolomics COSI #ismb2020 track to present a #Spec2Vec based novel #spectral #similarity #score! 😎



Lipids are also getting attention at the #CompMS #computational #metabolomics COSI #ismb2020 track by @Ahrends15 who introduces LipidCreator! 😎

Join us at #ISMB2022 #CompMS to learn about the latest advances in computational mass spectrometry. iscb.org/ismb2022 Keynote speakers: @Ge_Lab_UW, @briansearle, @Smith_Chem_Wisc, @gjpattij. Abstract submission deadline: April 21, 2022 ! iscb.org/ismb2022-submi…

Computational #massSpec job going in Berlin. #CompMS bsky.app/profile/drmuth…
It took just 11 minutes to parse 57 million lines of text into 1.1 million MS/MS spectra from my big .msp library and filter it with matchms. Impressive. #CompMS
Oh, amazing. Thank you so much. So nice to find a new #CompMS follow too. 😁
Hi, @jjjvanderhooft. Do you know an easy way to predict formula from MS1 isotope distribution in Python? I've tried running MS-FINDER from the command line but it's quite complicated. There must be an easier way. #CompMS
New @mzmine_project version 4.3: improved memory management, #MS2Deepscore molecular networking, fragment dashboard, MSConvert support. #metabolomics #CompMS #lipidomics Download now: github.com/mzmine/mzmine/… More information & Video: linkedin.com/posts/mzio_mzm…
@corinnabrungs and I left the @mzmine_project Workshop in Seoul with an amazing experience of the vibrant local #MassSpectrometry #CompMS #Metabolomics research communities. Thanks to 200+ for active participation and special thanks to @kyobinkang and the local organizing team.

@corinnabrungs and I left the @mzmine_project Workshop in Seoul with an amazing experience of how vibrant the local #massspectrometry #CompMS #metabolomics research communities are! Thanks to >200 for active participation and to @kyobinkang and the other local organizers.

Register now for our @mzmine_project #CompMS workshop hosted by @kyobinkang: July 9th, 10 am, Gemma Hall (Changhak B107), @sookmyung1906 University, Seoul, Korea. @corinnabrungs & @rschmid1789 will introduce non-target #MassSpectrometry data analysis. forms.gle/5P16q6dqeDsMtt…
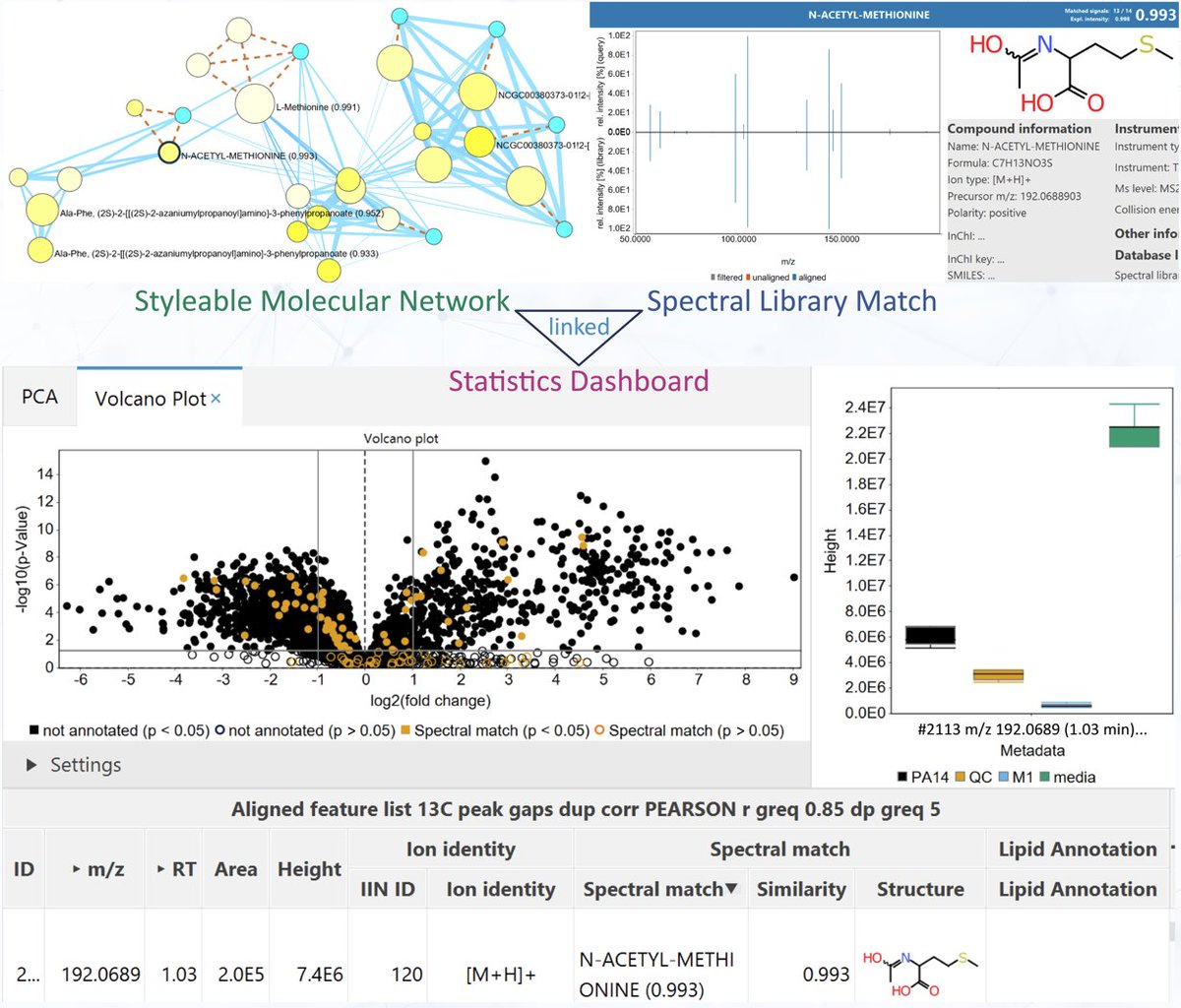
Our #compMS workshop at @OISTedu was very well received. Thanks to @rschmid1789 @corinnabrungs @roman_bushuiev @damiani_tito @LF_Nothias @boeckerlab!


Working with @HeuSteffen on the latest #ionmobility #CompMS #metabolomics methods is a blast - dynamic full-stack development with idea ping pong. Meet him at #asms2024. #proudPI
Thanks @EMN_MetSoc for highlighting my research! See you all at @MetabolomicsSoc #MetSoc2024 conference in Osaka, Japan. Or during our @mzmine_project workshop at @OISTedu on Okinawa. Reach out to me if you want to join our effort on open mass spectral libraries and #CompMS.
Introducing Robin for #ECRVoices @rschmid1789 is working on #MassSpectrometry and he is based in Prague, Czechia 🇨🇿

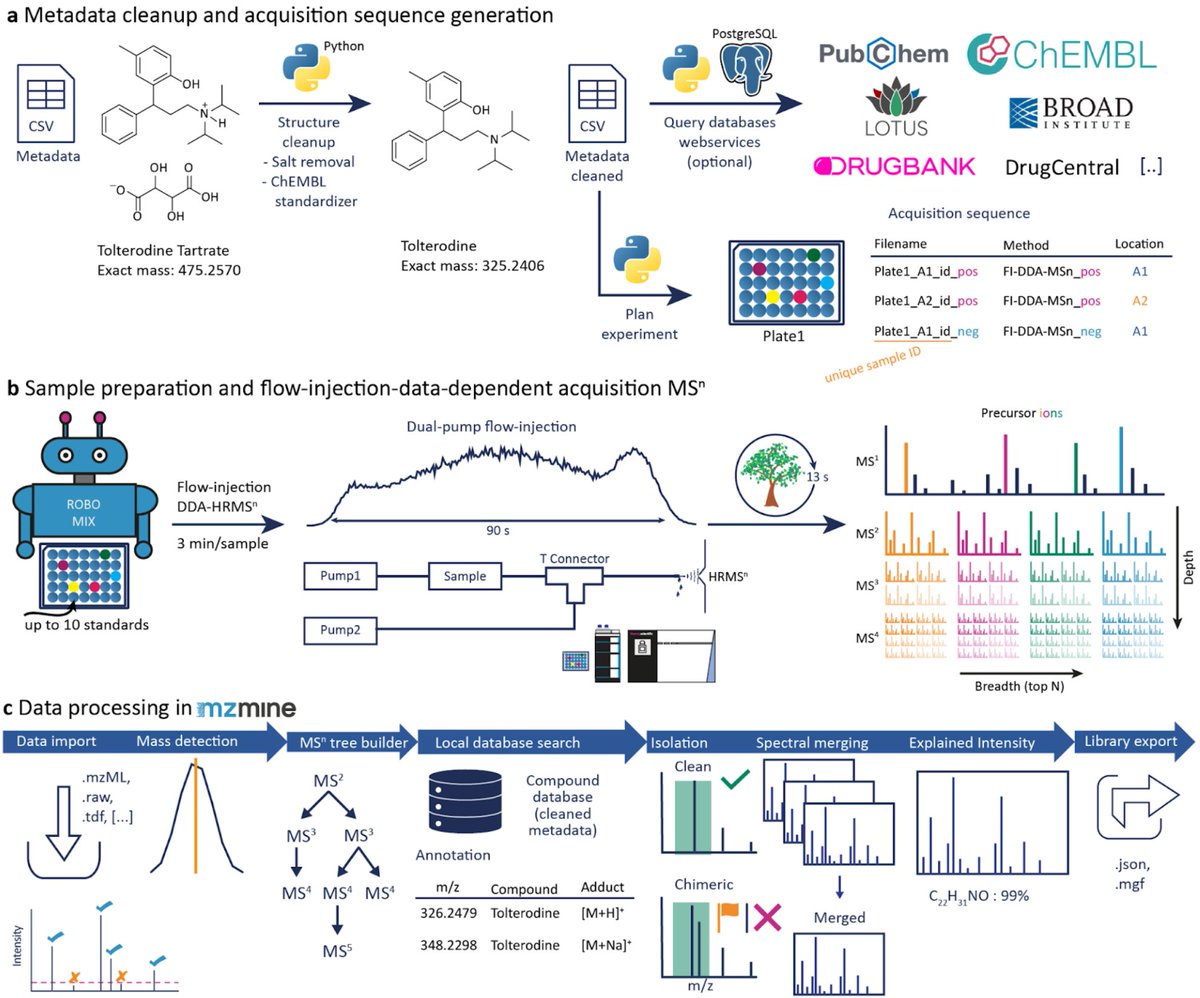
Our preprint describes an MSn library, a high-throughput acquisition method, and a #CompMS pipeline powered by @mzmine_project. Led by @corinnabrungs et al. we built the first #FAIR multi-stage fragmentation spectral tree library for >16,000 compounds. 1/n doi.org/10.26434/chemr…
Last week's @dagstuhl meeting on computational #metabolomics and #CompMS was a fantastic networking experience. The open-space format of this meeting allows for some dynamic discussions. Outcome: We formed task groups and one of them is working on mass spectral libraries.
About to board my train to Frankfurt... First lap on my way to @dagstuhl to discuss the latest in #CompMS and #metabolomics #lipidomics with old and new friends!

Enjoyed the event and thanks for having me. Here are my slides about early history of #MZmine, which started @BioscienceTurku: zenodo.org/records/110041… Just to acknowledge the main funder of the initiation of @mzmine_project : @SuomenAkatemia. #CompMS is a big field now.
Our @mzmine_project team celebrated the 20 year anniversary at the Prague #compMS workshop 2024. Fantastic to come together and plan the next 20 years with @HeuSteffen, @tomas_pluskal, @AnsgarKorf, @ElenaMokshina, Roman, @corinnabrungs, @matejoresic, @Adafede, and many more. 3/n

Our @mzmine_project team celebrated the 20 year anniversary at the Prague #compMS workshop 2024. Fantastic to come together and plan the next 20 years with @HeuSteffen, @tomas_pluskal, @AnsgarKorf, @ElenaMokshina, Roman, @corinnabrungs, @matejoresic, @Adafede, and many more. 3/n

Our Prague #compMS Workshop hosted 100 researchers +online. It was a great success thanks to all presenters, participants, sponsors, and venue @IOCBPrague. @mzmine_project #SIRIUS_MS #matchms @wikidata @TheLOTUSInitia1 @GNPS_UCSD #massspec #metabolomics 1/n ...


Fantastic session today with @LF_Nothias, diving into the powerful use of Cytoscape for network visualization. A great way to integrate all the tools from the workshop! #compMS @mzmine_project
Really cool session of introduction to matchms Python package and Jupyter notebooks itself. Very easy to follow the lesson with this well-written google colab notebook. This is a really powerful package for playing with msms data! @mzmine_project #compMS @vdHooft_CompMet

It’s time! @iscb‘s ISMB2020 is starting tomorrow! I’ve got a poster and brief presentation (#552) in the #computational mass spec track 🤓 as do many other members of the @mlaval6 lab 🔥 #compMS #ISMB2020

Today's the last day of @iscb's virtual flagship conference - it's been a wonderful experience exploring the computational work everyone has shared at #ISMB2020! I'll be "by my poster" in the #CompMS COSI this morning 🤓 #proteomics #computational #MassSpec #MachineLearning

Right now in the CompMS cosi at ISMB is "software solutions for multi-omic research" by Evgenia Shiskova from university of Wisconsin #ISMB18 #CompMS

Back after an exciting #CompMS seminar in @dagstuhl. Txs to all participants and the Dagstuhl staff!


Our #compMS workshop at @OISTedu was very well received. Thanks to @rschmid1789 @corinnabrungs @roman_bushuiev @damiani_tito @LF_Nothias @boeckerlab!


Last keynote of #CompMS COSI at #ISMB2018 by Josh Elias on some cool de novo search stuff (TagGraph)...

We're kicking off the #CompMS session at the #ismbeccb2021 conference with a keynote presentation by @thalexandrov on how computational MS enables spatial and single-cell metabolomics. Ready for two days of #CompMS goodness!

TagGraph: Match de novo peptides to sequence databases. Fast! ✨String-based ✨No precursor assumptions ✨No digestion assumptions ✨No PTM assumptions ✨FDR estimation #ISMB18 #CompMS #massspec #proteomics #PTMs #denovosequencing #databasesearch


Hannes Röst opening the #CompMS COSI track at #ISMBECCB 2019 in Basel with a talk on DIA data analysis


This workshop at @IOCBPrague is going to be a great opportunity to learn about the latest developments in @mzmine_project and other #compMS tools!

Register now! Prague Computational Mass Spectrometry Workshop, April 15-17, 2024, Prague, Czechia. Learn from the developers and power users of @mzmine_project, SIRIUS, #matchms, @TheLOTUSInitia1, @GNPS_UCSD and discuss how we can move the field forward... tinyurl.com/PragueCompMS20…
Final day of #ISMBECCB2023 came with the wonderful invited keynote from Bernhard @RenardLab on his journey with spectra and their current and outlook! #CompMS




@sneumannoffice presenting the R #metabolomics world at the #CompMS #computational #metabolomics COSI #ismb2020 track!


Really cool session of introduction to matchms Python package and Jupyter notebooks itself. Very easy to follow the lesson with this well-written google colab notebook. This is a really powerful package for playing with msms data! @mzmine_project #compMS @vdHooft_CompMet

Look who I found at #ISMB18 #CompMS! @StanfordMed @StanfordBiosci @ChemSysBio @iscb @TheGonz79 @Josh_Lichtman #EliasLabAlumni #proteomics #massspectrometry #omics #denovosequencing

Ready for the Prague Computational Mass Spectrometry Workshop at the @IOCBPrague. Thanks to the organizers @mzmine_project @tomas_pluskal @rschmid1789 and others #compMS




Join us at #ISMB2022 #CompMS to learn about the latest advances in computational mass spectrometry. iscb.org/ismb2022 Keynote speakers: @Ge_Lab_UW, @briansearle, @Smith_Chem_Wisc, @gjpattij. Abstract submission deadline: April 21, 2022 ! iscb.org/ismb2022-submi…

Our December Client Spotlight is on @UHHiloDKICP, who is using the CORE #ELMS and CORE #CompMS applications!

Something went wrong.
Something went wrong.
United States Trends
- 1. #UFC322 42.6K posts
- 2. Ewing 6,382 posts
- 3. Bama 19.8K posts
- 4. Arch Manning 1,879 posts
- 5. #AEWCollision 6,506 posts
- 6. Wellmaker 4,474 posts
- 7. Georgia 76.7K posts
- 8. Oklahoma 30.2K posts
- 9. Tracy Cortez 2,015 posts
- 10. James Peoples N/A
- 11. Noah Thomas N/A
- 12. Wingo 1,488 posts
- 13. Bronny 5,877 posts
- 14. Jeremiah Smith 2,393 posts
- 15. Lebby N/A
- 16. Ole Miss 7,253 posts
- 17. Ty Simpson 3,913 posts
- 18. Erin Blanchfield 1,087 posts
- 19. #Svengoolie 1,436 posts
- 20. Shapen N/A