#metatranscriptomics результаты поиска
New method alert!🚨We're thrilled to present #Spatial #metaTranscriptomics (SmT) to detect #host-#bacteria-#fungi #interactomes in tissues. Collaboration between @sami_saar in my lab and @orshalevsk, @Haim_Ashkenazy, @DerekSeveri, Vanessa in @PlantEvolution lab.Main findings in🧵

Today @thaisegreja gave an overview of her research on Maize-Xanthomanas patho-biology and #metatranscriptomics to our @UNL_PSI @UNLresearch @UNLPlantPath community!

Interested in #Metagenomics, #metatranscriptomics, and #multiOmics🦠🦠? Do not miss the opportunity to join us in May for the 5th edition of this great course! Still few available places: physalia-courses.org/courses-worksh… #Genomics #Bioinformatics #microbiome
We have the last 2 seats available in the #Metagenomics, #metatranscriptomics, and #multiOmics courses 🦠🦠 If interested, please check it out: physalia-courses.org/courses-worksh… #Genomics #Bioinformatics #microbiome

This is pretty good for the people who are not yet long reads wizards - @lizilla93 #mewe2019 #metagenomics #metatranscriptomics
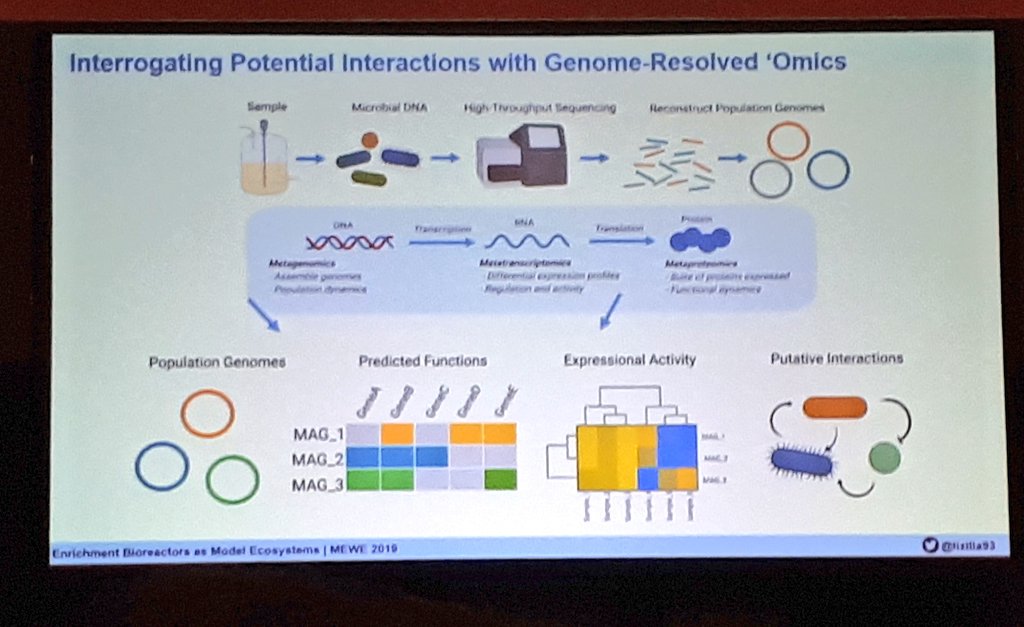
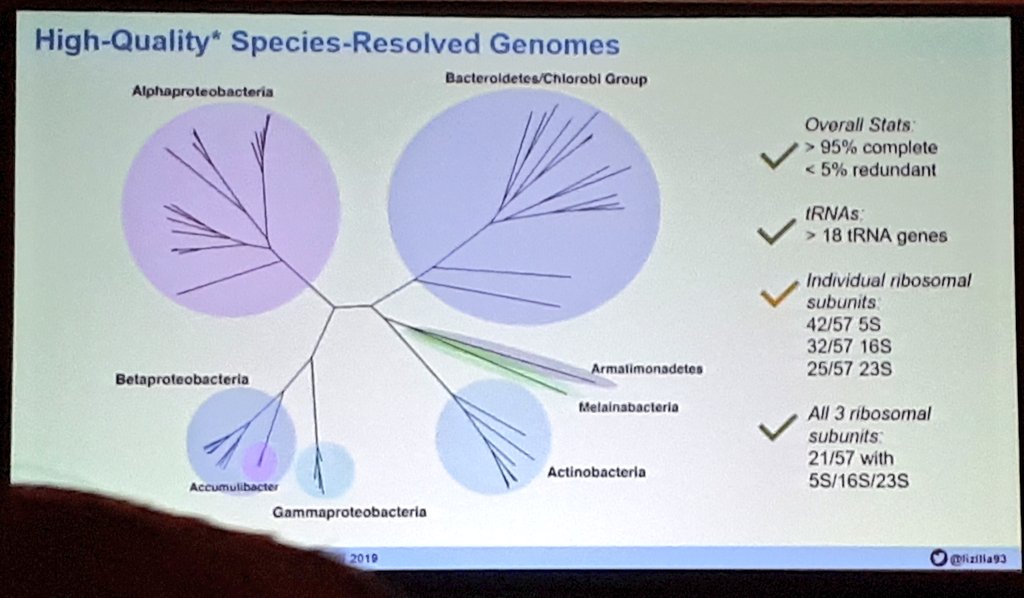
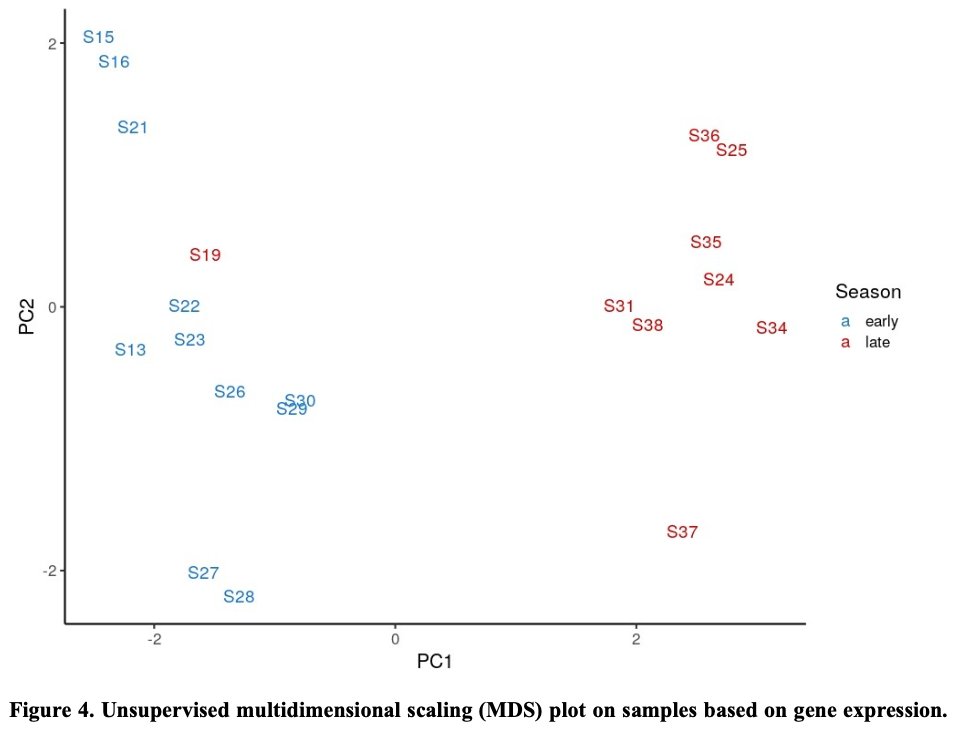
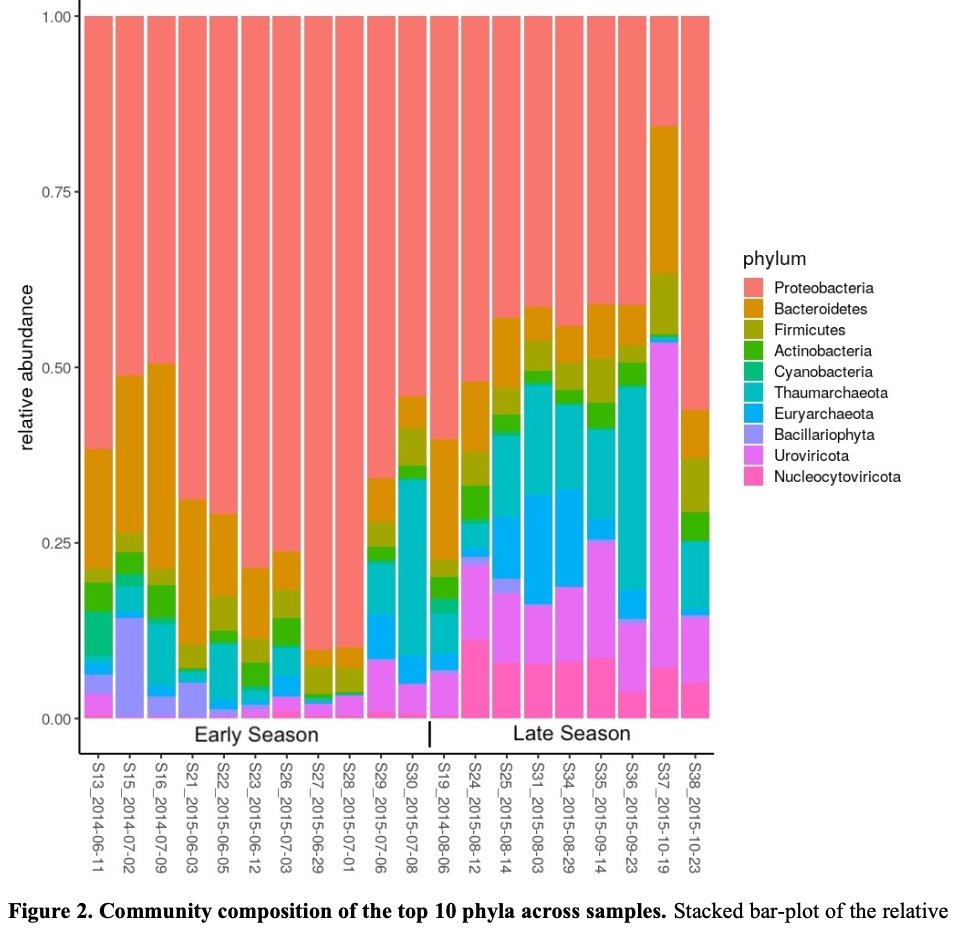
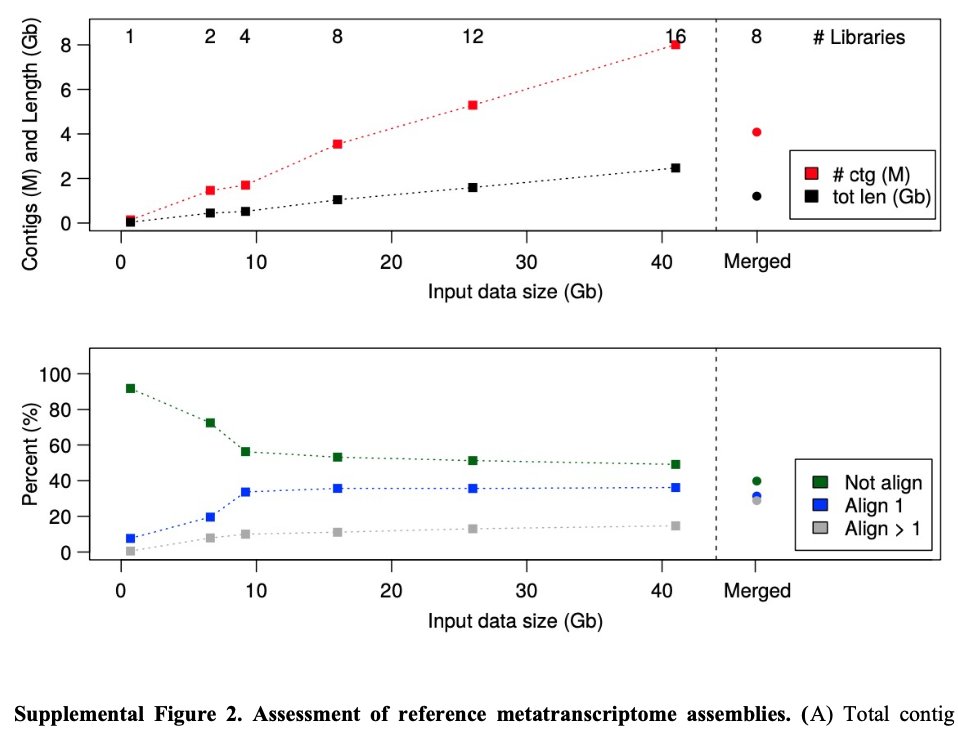
Our latest #preprint, "#Metatranscriptomics reveals a shift in microbial community composition and function during summer months in a coastal marine environment", on @biorxivpreprint. Thanks for checking it out! Great to work with this outstanding team. biorxiv.org/content/10.110…
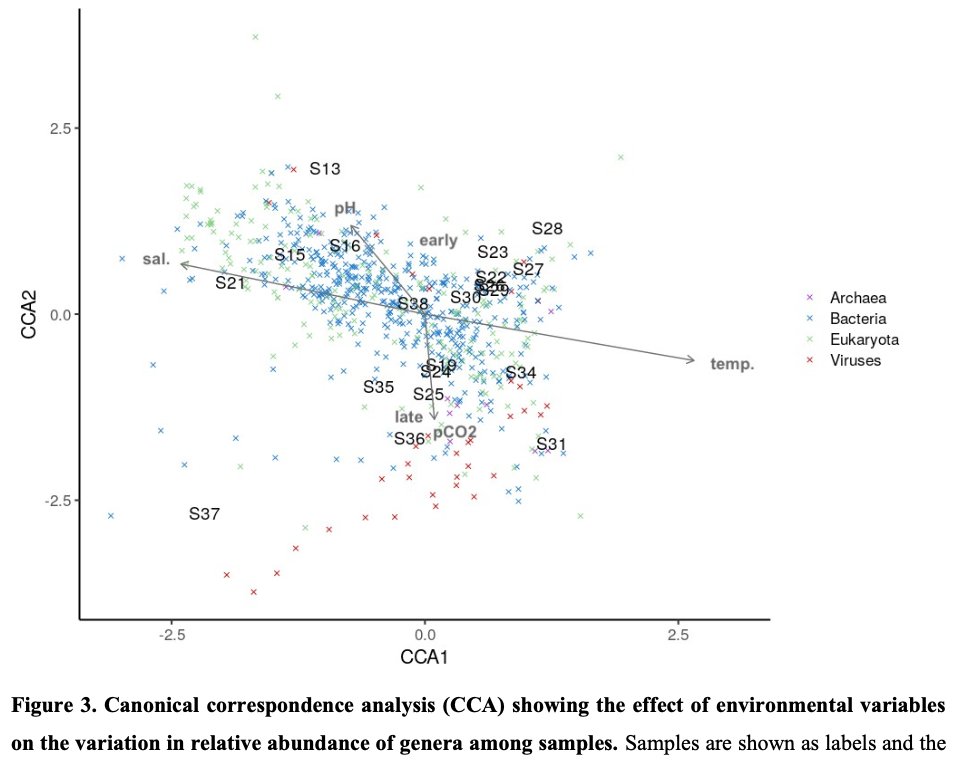
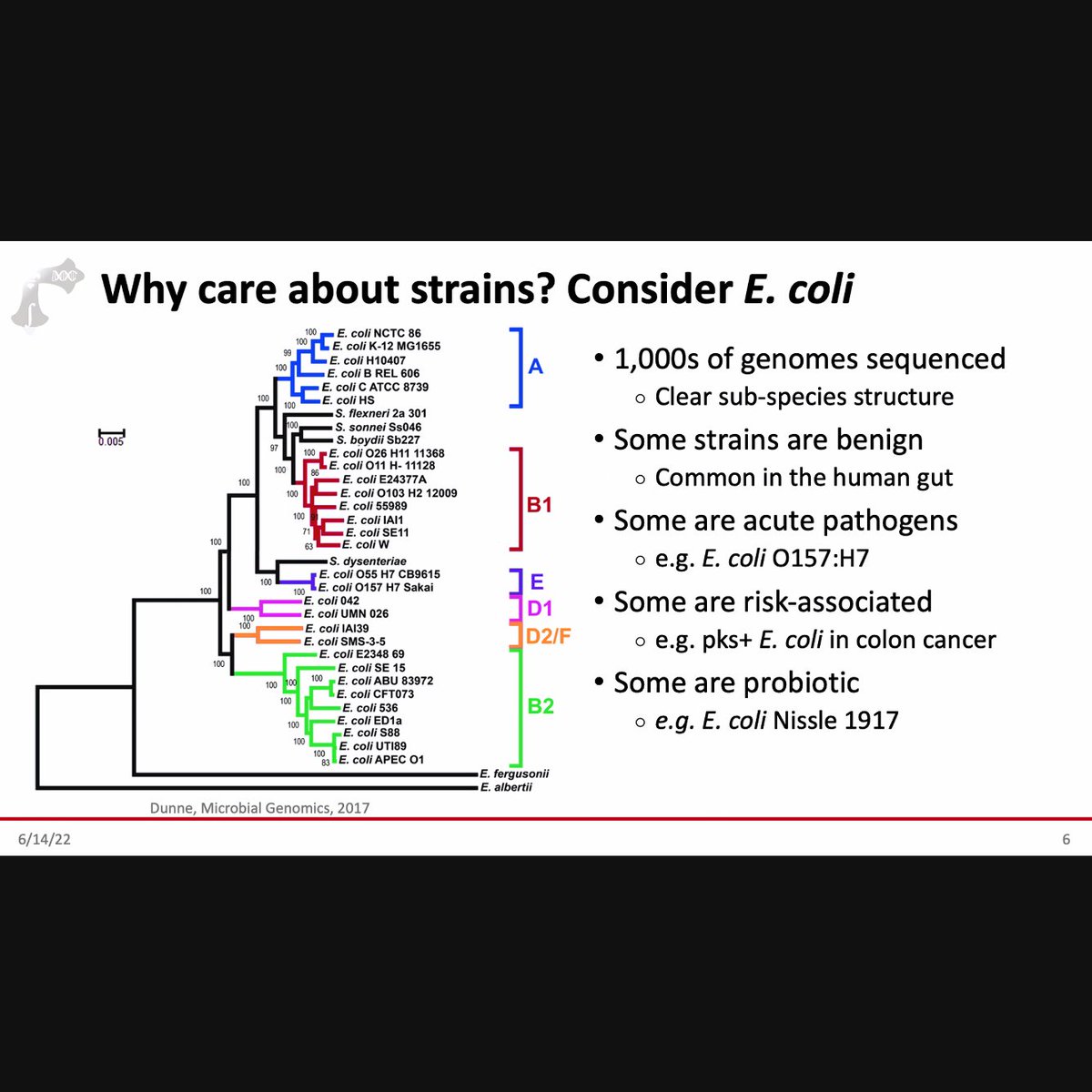
Learning about intra-species functional diversity and why it matters for human and ecosystem health 🦠🧑🤝🧑🌳 - fascinating & complex! via the @Physacourses #metagenomics / #metatranscriptomics course led by the Huttenhower lab.

Kicking off the 2nd edition of our #Metagenomics, #metatranscriptomics, and #multiomics for microbial community studies in collaboration with @chuttenh lab huttenhower.sph.harvard.edu #microbiome

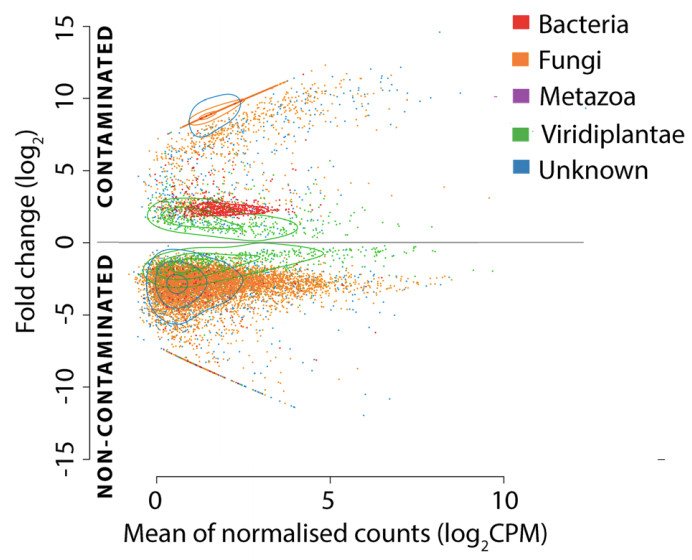
#metatranscriptomics of willow roots show shifts in plant gene expression but also in fungi and bacteria communities for trees grown in contaminated soils. Our latest #holobiont research with @yergeaue @fredericpitre @MichelPhyto @NJBBrereton @GenomeQuebec microbiomejournal.biomedcentral.com/articles/10.11…
Great lecture by @efranzosa in this day4 of the #MultiOmics #Metagenomics course about #metatranscriptomics and how to profile the functional activity of microbial communities #Metagenomics #MultiOmics @hutlab #Bioinformatics
More cool stuff from the Sunny Coast 😎 @Dr_ErinPrice Use of #metatranscriptomics to identify #pathogen diversity and their activity in #CF patients @DerekSarovich @usceduau @AUSSOCMIC #2019ASM

What's your go-to normalization method for #metatranscriptomics (TPM, RPKM, Deseq2, others)? Bonus points if you have a great way to normalize metaT to paired #metagenomics datasets! Any insights into mapping metaT to itself, paired metaG reference, other ref DB? Please RT!
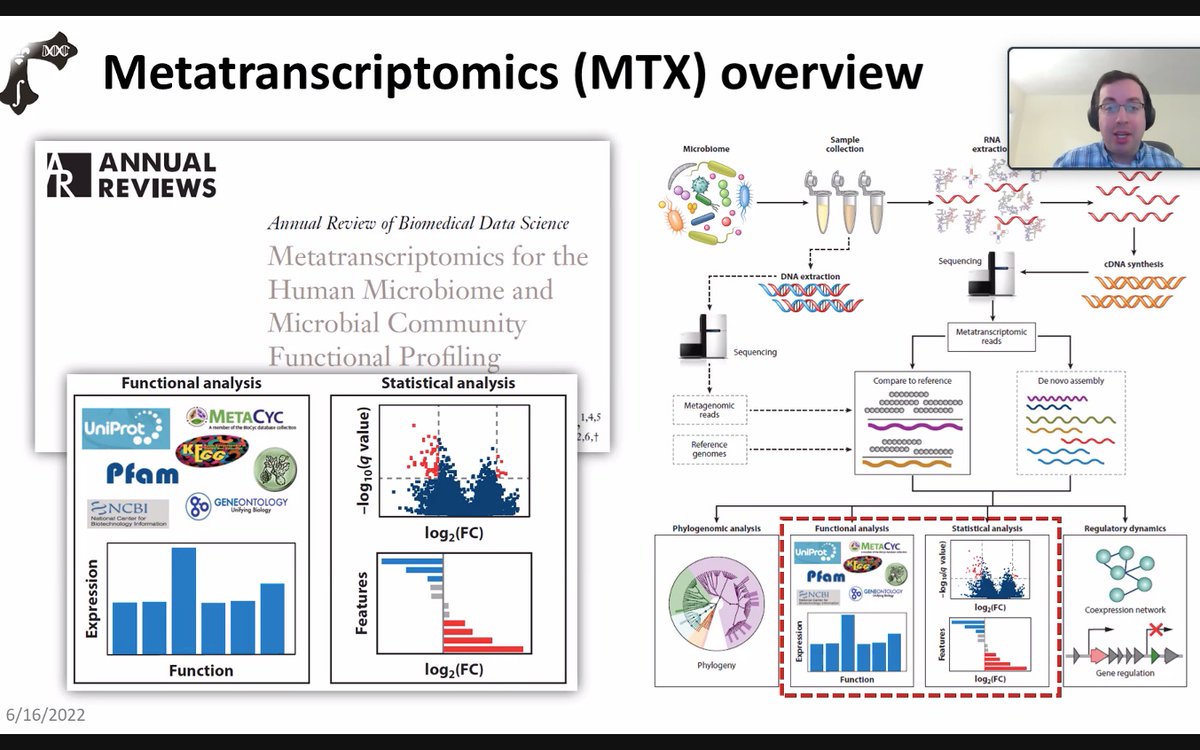
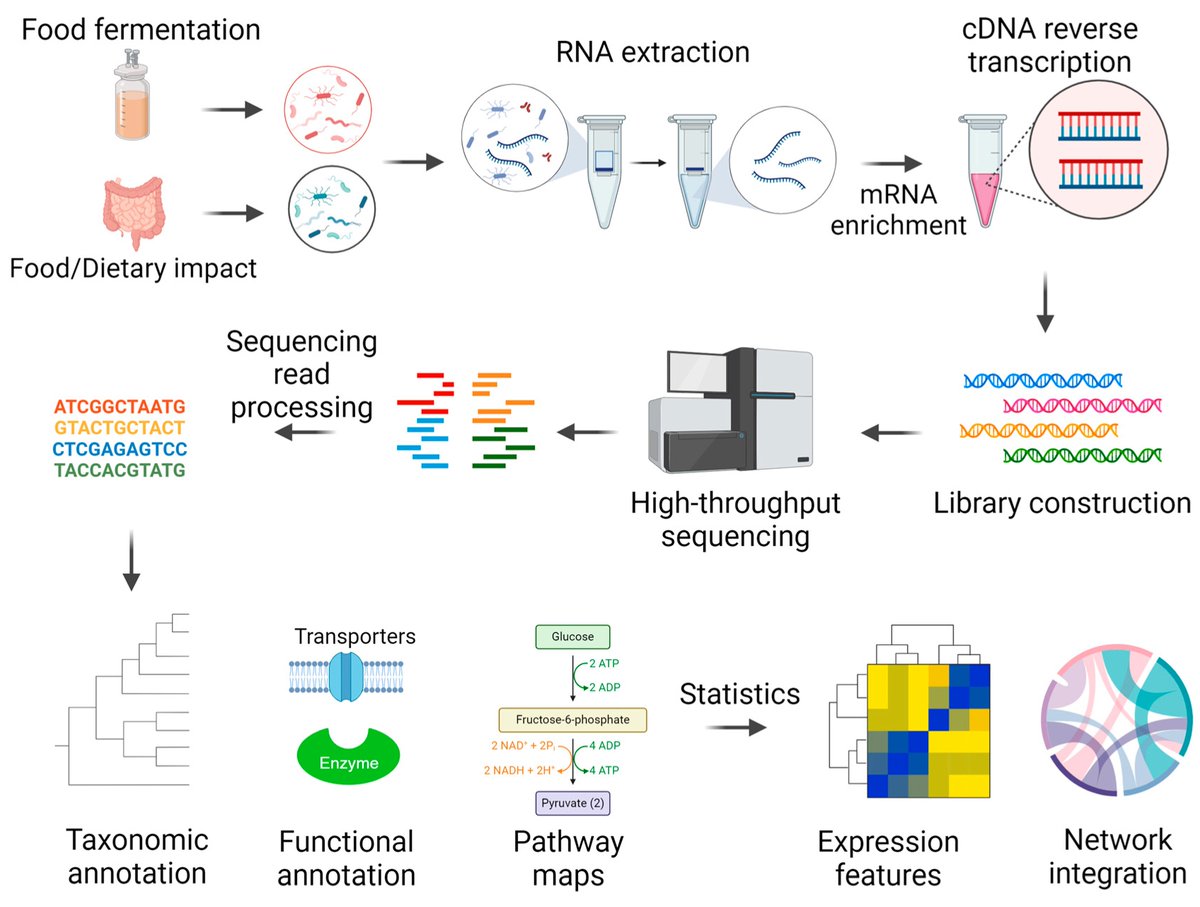
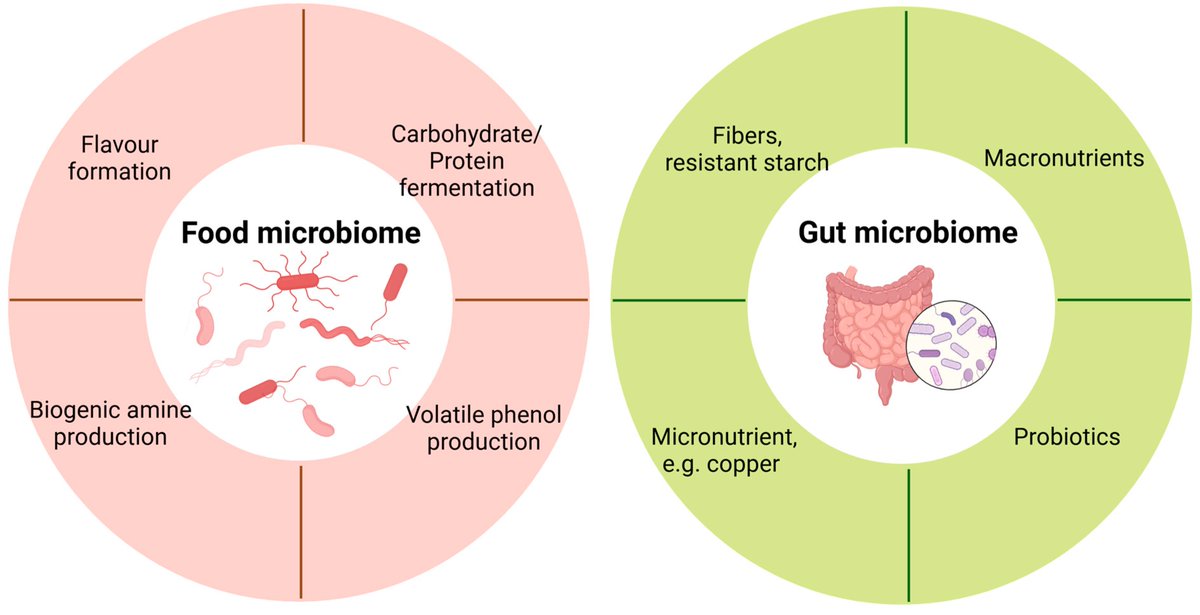
🌟 #LastestPaper 📖 #Metatranscriptomics for Understanding the #Microbiome in #Food and #Nutrition Science 🧑🏻🔬By Dr. Chunlong Mu, et al. 🔗mdpi.com/2218-1989/15/3…
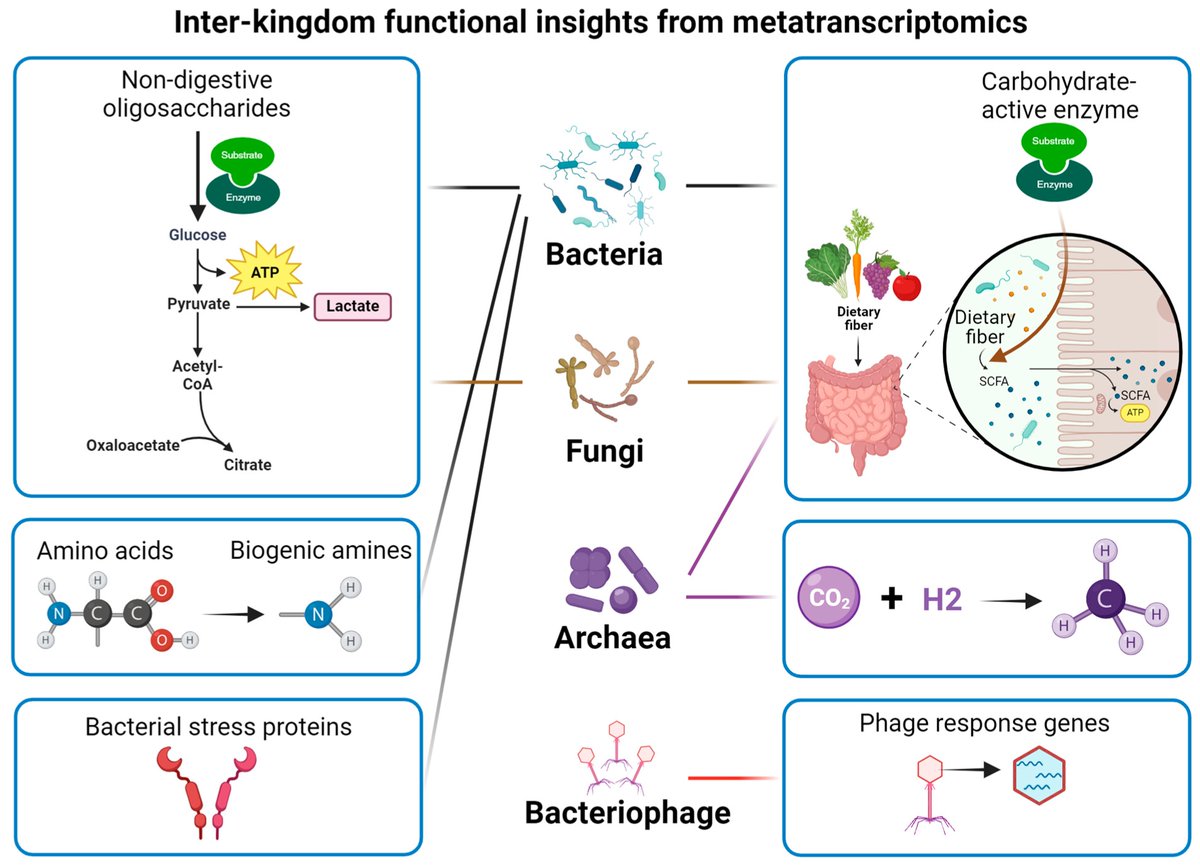
Do you want to learn #Metagenomics/#Metatranscriptomics/#Bioinformatics/#MultiOmics? Join us in June for this amazing course with @hutlab: 🔗physalia-courses.org/courses-worksh… There are still a few seats available! #Bioinformatics #genomics #sequencing #microbiome #metagenomics

Great talk by @Dr_ErinPrice, who discusses the advantages of using #metatranscriptomics for the analysis of active #polymicrobial infections in the #cysticfibrosis lung! @usceduau @AUSSOCMIC

Word Cloud for #metatranscriptomics from the participants on the third day of the #microbiome #usegalaxy workshop organized by @usegalaxyp & @CSIR_IMTECH. Thanks to @bebatut @pratikomics @subina @shiltemann for Day 3 of the workshop! @ASMicrobiology @INDOUSSTF
Very excited for #2021ASM Stream 1 (starting soon!!) and later Stream 3 talks, featuring some of our uber unique uplifting @usceduau #microbiology peeps @Dr_ErinPrice @KasimovVasilli Susy Anstey #metatranscriptomics #respiratorydisease #pathogenomics #chlamydia #birds #horses
Last 5 seats left on the ONLINE @Physacourses "#Metagenomics, #metatranscriptomics, and multi'#omics for microbial community studies" with @hutlab in June. If interested, please see: physalia-courses.org/courses-worksh… #microbiome Please RT

Out now in #NucleicAcidsResearch: my first PhD 🎓 chapter! If you're interested in target-PCR-free🧬taxonomic identifications 🔎of microbes🦠, specifically #metagenomics and #metatranscriptomics, this one is for you🫵! academic.oup.com/nar/advance-ar…
Alternatives Integrating Omics Approaches for the Advancement of Human Skin Models: A Focus on #Metagenomics, #Metatranscriptomics, and #Metaproteomics 💡📖 Full Text → mdpi.com/3425500 #OmicsApproaches #SkinModels #Proteomics #HumanHealth #Microbiome
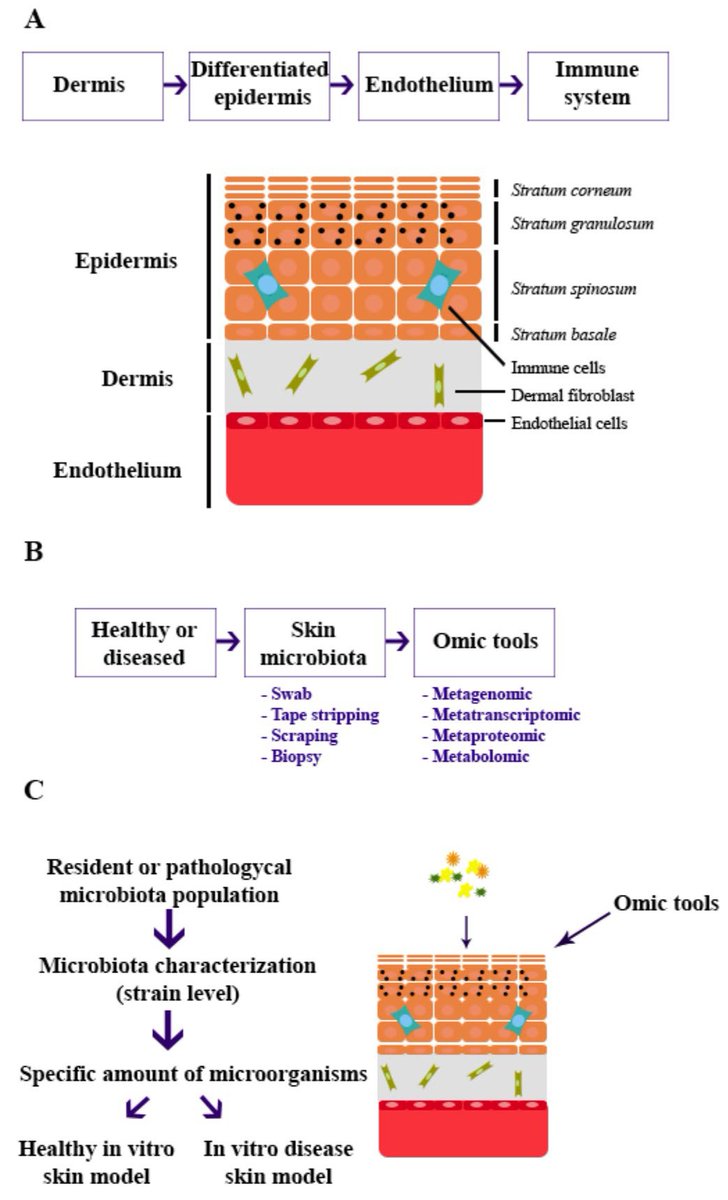
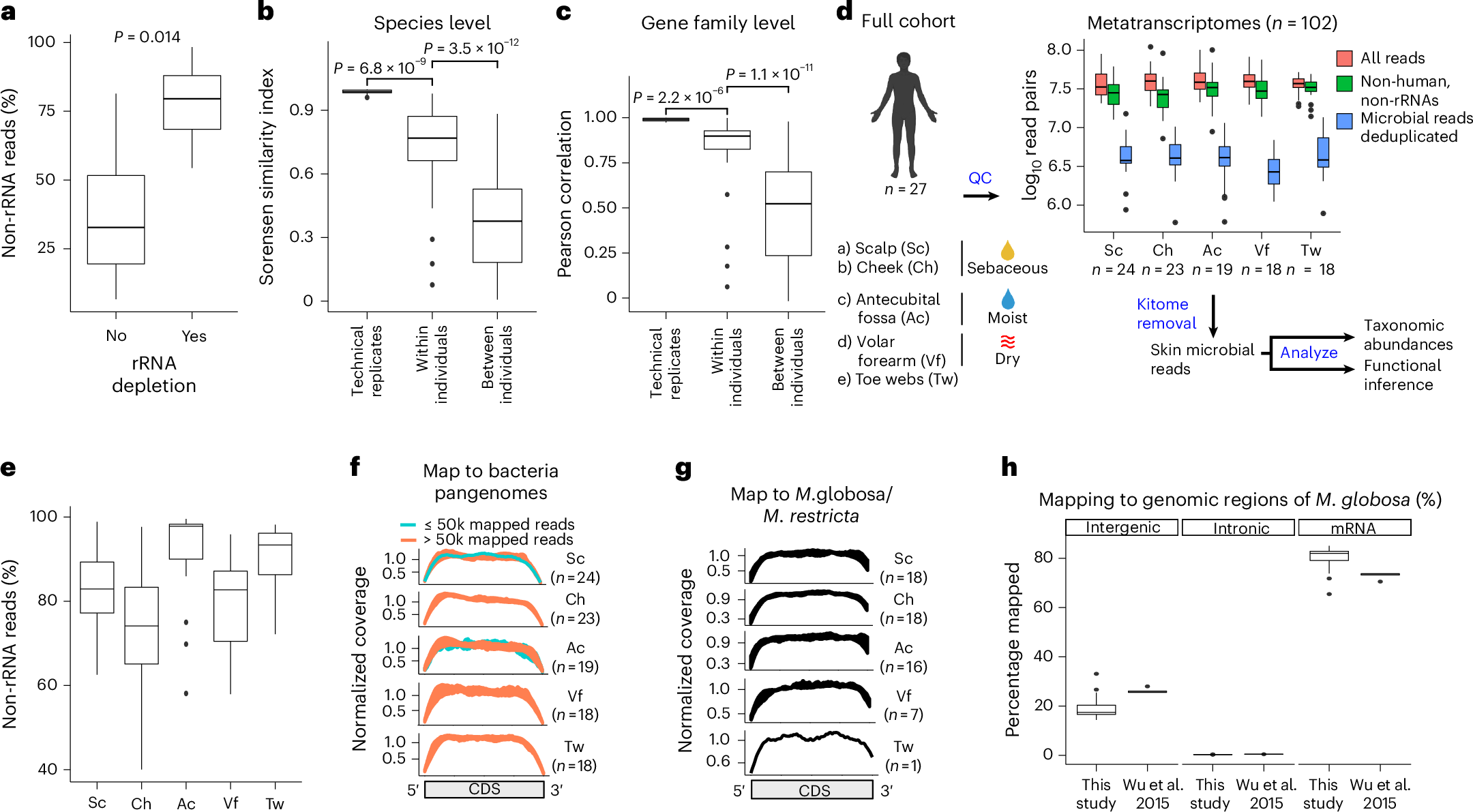
Skin #metatranscriptomics could be transformational in a way not seen in gut/oral studies as 1) Defined sampling can provide in vivo relevant insights 2) Microbial mRNA is dominant, but human mRNA is also detected nature.com/articles/s4158…
Learn about #metagenomics, #metabarcoding, #metatranscriptomics, and #genome data analysis Register: shorturl.at/wq6u9

🚨 Chikungunya detection breakthrough! Metatranscriptomic sequencing uncovers low viral-load infections—advancing biosafety and global health. 🔬 Learn more: sciencedirect.com/science/articl… #Biosafety #HealthInnovation #Metatranscriptomics #VirusDetection

Out in ME! A new study led by @RebeccaKFrench uses total #RNAseq and #metatranscriptomics to reveal associations between extraintestinal pathogenic E. coli and exudative cloacitis in the critically endangered #kakapo. @WileyEcolEvol 🔗: buff.ly/3HxZUW3 📷: Lydia Uddstrom

Functional ‘omics #metatranscriptomics, #metaproteomics, and #metabolomics identify functional traits of the microbiome that are linked to host health and disease. gastrojournal.org/article/S0016-…

🌟 #LastestPaper 📖 #Metatranscriptomics for Understanding the #Microbiome in #Food and #Nutrition Science 🧑🏻🔬By Dr. Chunlong Mu, et al. 🔗mdpi.com/2218-1989/15/3…



First preprint in 2025: #Metatranscriptomics sheds light on who is doing what in a #coral Great collaboration led by @kt_microbes @heroenv @LabVerbruggen @deepseacrepture @momedinamunoz @arc_gov_au @MooreFound biorxiv.org/content/10.110…
🧬 Excited to hear from Imadh Abdul Azeez! He used #metagenomics and #metatranscriptomics to assess the impact of zinc supplementation on the gut microbiome of children in Bangladesh. Discover insights from the ZiPT trial and its implications for child #GutHealth #MicroSeq2024


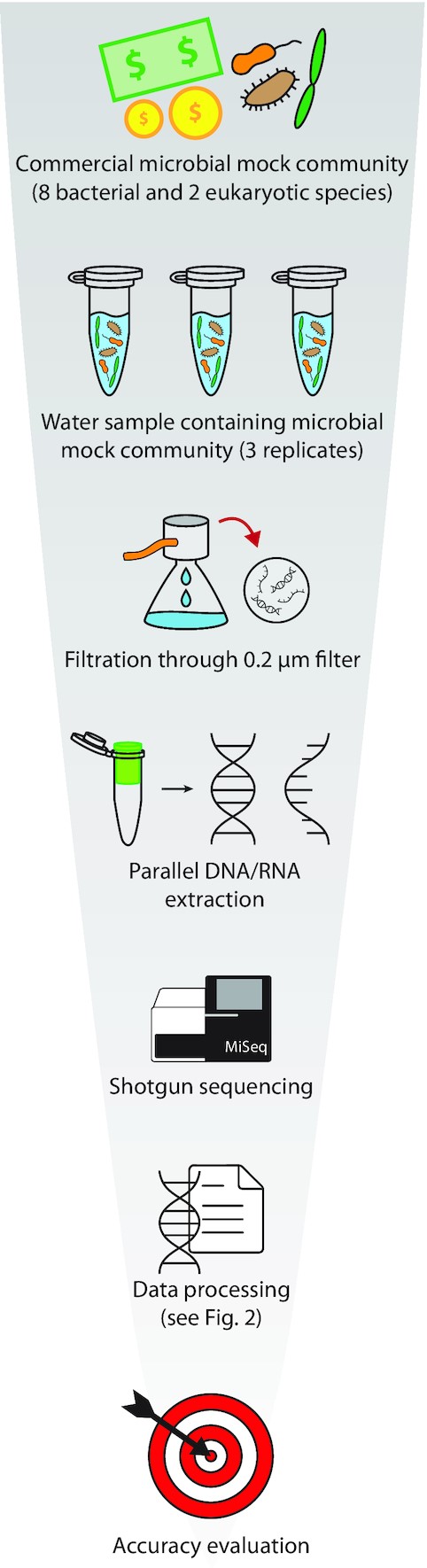
#Friday #PaperforWeekend #metatranscriptomics A metatranscriptomics strategy for efficient characterization of the microbiome in human tissues with low microbial biomass ncbi.nlm.nih.gov/pmc/articles/P…
#Thusday #Metatranscriptomics #HostDepletion Rational probe design for efficient rRNA depletion and improved metatranscriptomic analysis of human microbiomes bmcmicrobiol.biomedcentral.com/articles/10.11…
#Wednesday #Metatranscriptomics Metagenomics versus total RNA sequencing: most accurate data-processing tools, microbial identification accuracy and perspectives for ecological assessments academic.oup.com/nar/article/50…
#Tuesday #metatranscriptomics Current concepts, advances, and challenges in deciphering the human microbiota with metatranscriptomics pubmed.ncbi.nlm.nih.gov/37365103/
Interesting seminar and flash talks organised by @NCCTuebingen on applications and workflows of #metagenomics #metatranscriptomics #genomicsenthusiats

#Trichodesmium colonies living in the same water mass experience different microenvironments. Combining single-colony parallel #metagenomics and #metatranscriptomics we show that each colony expresses general pathways alongside ones specific to the microenvironment.
Current concepts, advances, and challenges in deciphering the human #microbiota with #metatranscriptomics sciencedirect.com/science/articl…
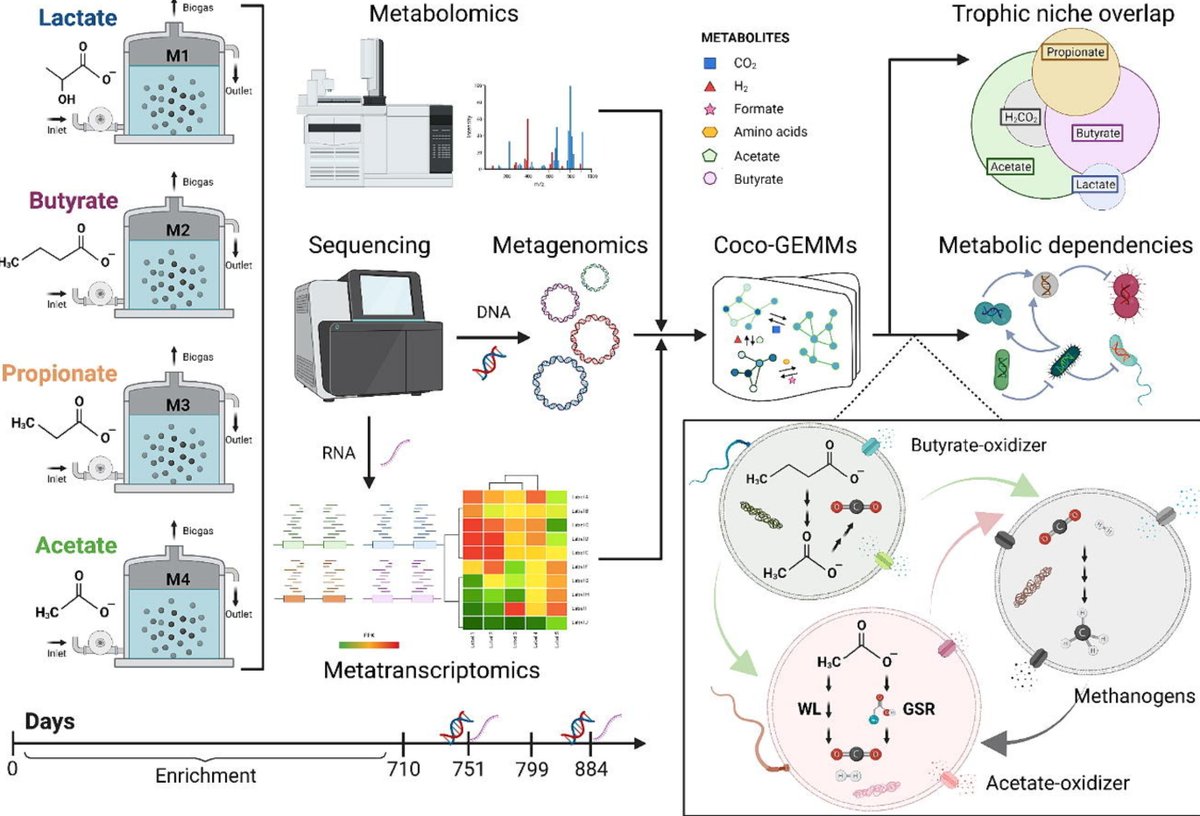
Our paper is out in @Chem_Eng_J - a combined approach including #metagenomics,#metatranscriptomics,#machine-learning and #metabolic modelling to decipher the #syntrophies in C2-C4 organic acids-degrading #anaerobic #microbiomes– doi.org/10.1016/j.cej.… @DiBio_UniPD @IBBPAS
New method alert!🚨We're thrilled to present #Spatial #metaTranscriptomics (SmT) to detect #host-#bacteria-#fungi #interactomes in tissues. Collaboration between @sami_saar in my lab and @orshalevsk, @Haim_Ashkenazy, @DerekSeveri, Vanessa in @PlantEvolution lab.Main findings in🧵

We have just started our #Metagenomics, #metatranscriptomics, and #multiOmics Workshop with Melanie Schirmer, @Kevbonham, Cesar Arze and @chuttenh #ksgut #KSmicrobiome


Our paper: #Metatranscriptomics of #Biogeochemically-relaed Genes in #Amazon River, @PLOSONE dx.doi.org/10.1371/journa…

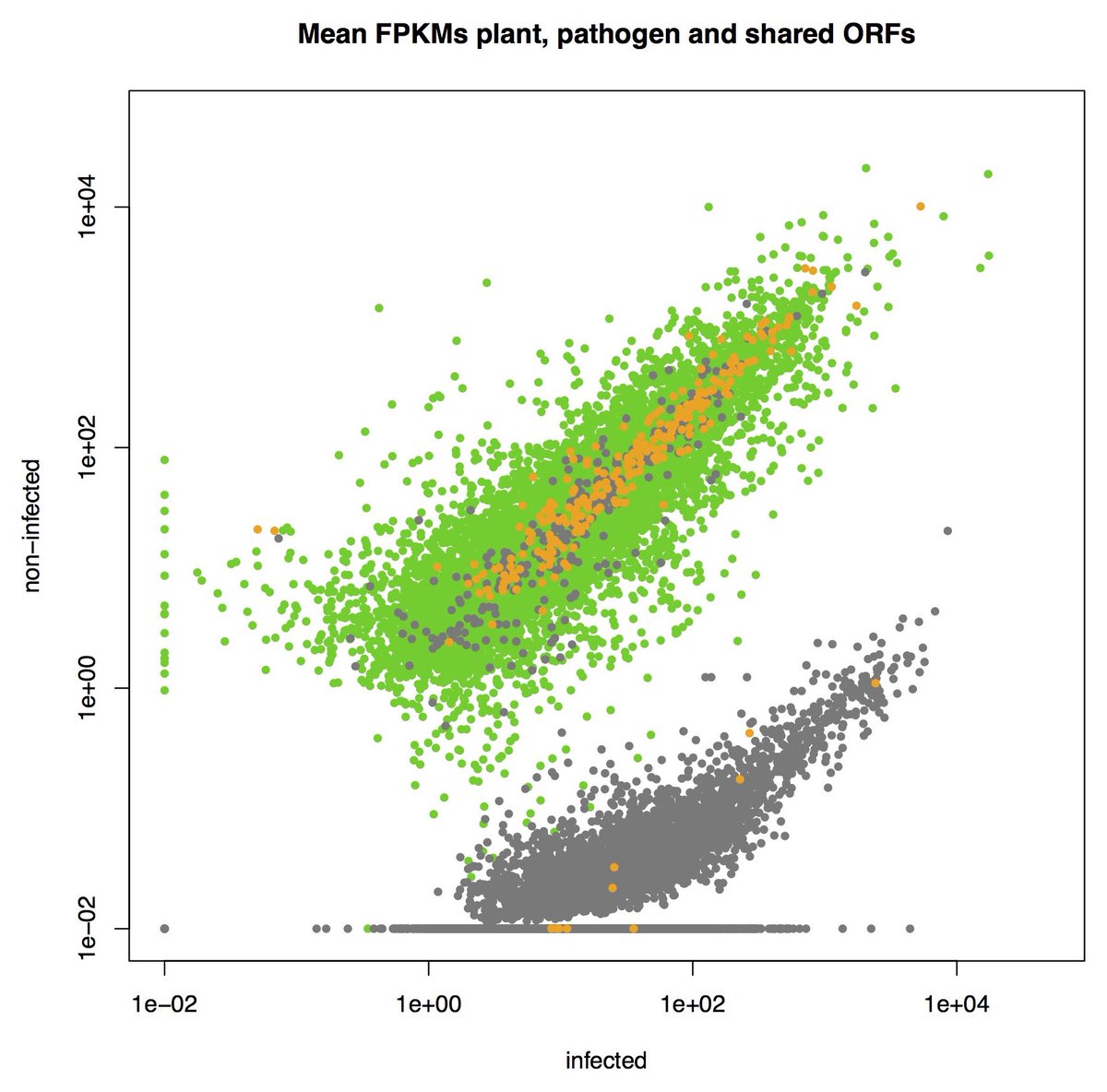
Separating host plant and pathogen ORFs without reference genomes #PlantPathogenomics #FieldPathogenomics #metatranscriptomics
Interested in #Metagenomics, #metatranscriptomics, and #multiOmics🦠🦠? Do not miss the opportunity to join us in May for the 5th edition of this great course! Still few available places: physalia-courses.org/courses-worksh… #Genomics #Bioinformatics #microbiome

Today @thaisegreja gave an overview of her research on Maize-Xanthomanas patho-biology and #metatranscriptomics to our @UNL_PSI @UNLresearch @UNLPlantPath community!

@pratikomics presents talk on #metaomics (#metagenomics #metatranscriptomics #metaproteomics #metametabolomics) tools and workflows in #microbiome research within #usegalaxy framework at the #awcls20 in @lorentzcenter at @UniLeidenNews
We have the last 2 seats available in the #Metagenomics, #metatranscriptomics, and #multiOmics courses 🦠🦠 If interested, please check it out: physalia-courses.org/courses-worksh… #Genomics #Bioinformatics #microbiome

Registrations are now closed for our course "#Metagenomics, #metatranscriptomics, and multi'omics for microbial community studies"

SAMSA v2.0 will allow in house annotations, run faster.... #goSam! #metatranscriptomics #BiotechRetreat

We used a novel computational 🖥️workflow leveraging #metatranscriptomics #metagenomics and #metabolomics 🧬from the #HMP2 to identify 12 candidate enzymes from 2 protein families that could perform this conversion. Neither had previously been implicated in drug metabolism. (4/9)
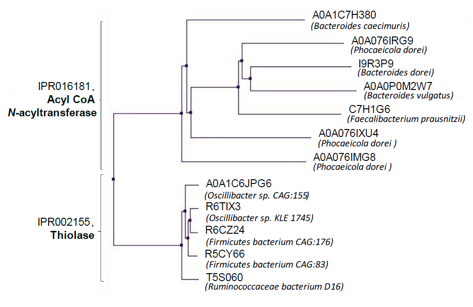
@Marie_a_crane from @usegalaxyp presented her summer project on #metatranscriptomics #asaim workflow in #usegalaxy. Merci @bebatut pour votre assistance !

New paper out enabling #metatranscriptomics integration in genome-scale metabolic models of microbial communities @guido_zampieri @campanarostef @LauraTreu @CellRepMethods doi.org/10.1016/j.crme…

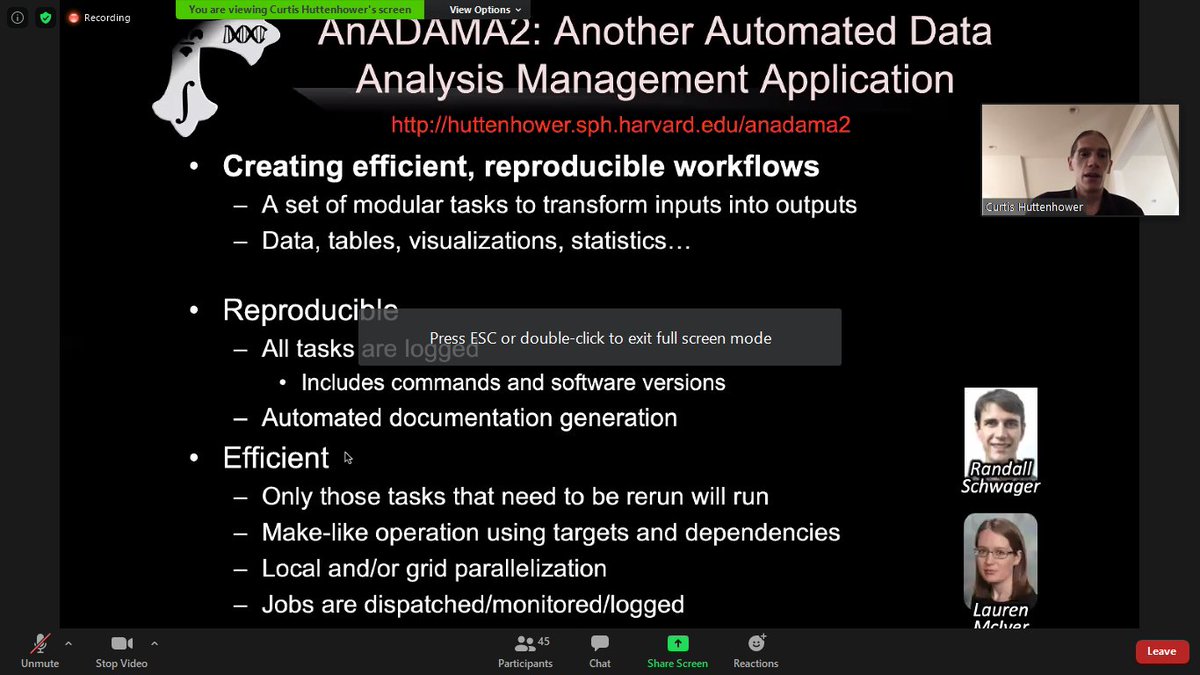
Curtis is now presenting their tool AnADAMA2 to create reproducible workflows and execute them efficiently. github.com/biobakery/biob… #omics #metagenomics #metatranscriptomics
Kicking off the 2nd edition of our #Metagenomics, #metatranscriptomics, and #multiomics for microbial community studies in collaboration with @chuttenh lab huttenhower.sph.harvard.edu #microbiome

Today’s seminar: @FR_LucasAuer on #Forest #soil #metatranscriptomics: a long journey in #bioinformatics

Great talk by @Dr_ErinPrice, who discusses the advantages of using #metatranscriptomics for the analysis of active #polymicrobial infections in the #cysticfibrosis lung! @usceduau @AUSSOCMIC

Gaia is now on @omictools! Check it out at: omictools.com/gaia-tool for all your #amplicon #wholegenome and #metatranscriptomics analysis. #Genefunction coming soon. #Metagenomics made easy. #SUPERGaia #QueenOfMetagenomics
Do you want to learn #Metagenomics/#Metatranscriptomics/#Bioinformatics/#MultiOmics? Join us in June for this amazing course with @hutlab: 🔗physalia-courses.org/courses-worksh… There are still a few seats available! #Bioinformatics #genomics #sequencing #microbiome #metagenomics

Something went wrong.
Something went wrong.
United States Trends
- 1. Syracuse 6,192 posts
- 2. Joe Jackson N/A
- 3. Harden 27.7K posts
- 4. Arch Manning 1,528 posts
- 5. #UFCQatar 70.8K posts
- 6. Jeremiyah Love 2,513 posts
- 7. Lincoln Riley N/A
- 8. Arman 21.4K posts
- 9. Arkansas 8,578 posts
- 10. Fran Brown N/A
- 11. Mercer 3,050 posts
- 12. Stoops 1,118 posts
- 13. Zvada N/A
- 14. Taylen Green N/A
- 15. Ian Garry 9,259 posts
- 16. Sherrone Moore N/A
- 17. #GoIrish 4,242 posts
- 18. Belal 10.2K posts
- 19. Kansas State 2,500 posts
- 20. Malik Benson N/A