#metatranscriptome 검색 결과
just re-calculated the depth of our published #metatranscriptome Covering transcripts from species at <1% abundance!

Congrats to our postdoc fellow @YanYan20444341 for awarding the Charles King Trust Fellowship! Yan will study the strain-level epidemiology and diet-linked gut #metatranscriptome risk factors in #colorectal cancer.

#BIO2015 World Fusion's Tokyo division busy in booth 3649 with LSKB and drag&drop #metagenome & #metatranscriptome

Evaluation of commercial RNA extraction kits for long-read metatranscriptomics in soil. #nanopore #metatranscriptome doi.org/10.1099/mgen.0…

@LauraTreu presenting the #genome-centric #metatranscriptome study on #Anaerobic #digestion system at #SIMGBM in Florence 2019! Great work! @DiBio_UniPD @IriniAngelidaki @kougias


Best place to upload a #metatranscriptome #assembly? (i.e., #RNAseq reads from environmental sample, assembled into a multiple-species assembly?) I figure this is useful for reproducibility, but possibly @NCBI is not the location? #eRNA ncbi.nlm.nih.gov/genbank/tsa/ Any ideas? Thanks!

Once we aligned the samples against the ref #metatranscriptome and normalized, based on a MDS plot it was clear that there was a strong signal across the first dimension. This indicated a strong effect of season, where early (June-July) and late (Aug-Oct) clustered separately...

A new item for me here was to try a few ways to assemble a ref #metatranscriptome. - incr. library inclusion step-by-step (1, 2, 4, 8, 12, 16) - assembling 8 libraries individually, then merge ...then we eval. the effects on length, contiguity, % align, and redundant align

Evaluation of commercial RNA extraction kits for long-read metatranscriptomics in soil. #nanopore #metatranscriptome doi.org/10.1099/mgen.0…

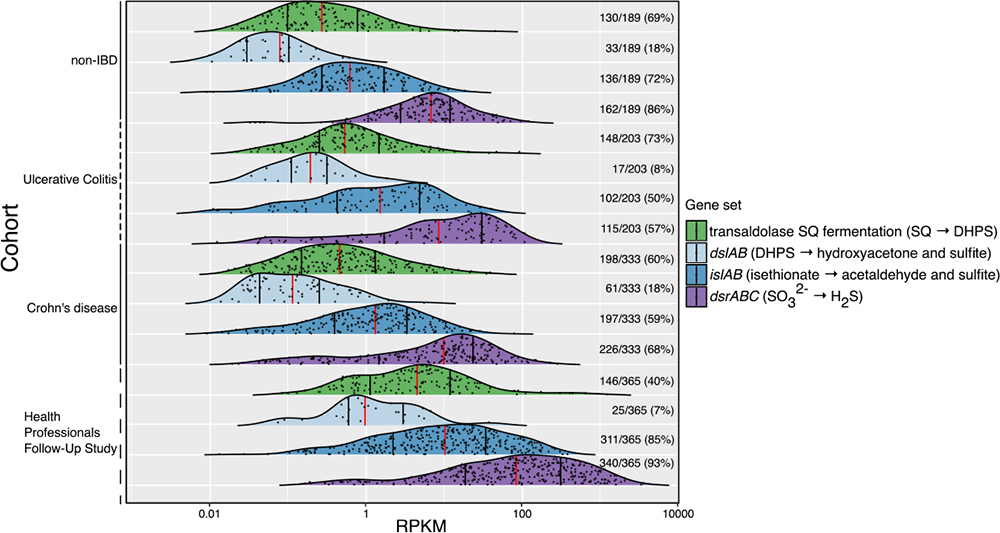
#metatranscriptome analysis of DsrABC-sulfite reduction and DHPS/isethionate sulfite-lyase activities suggested Bilophila and other Desulfovibrionaceae use mostly #taurine but also #DHPS for #H2S production in the human gut
Julian von Borries presenting an intriguing new gadget for your metatranscriptome analysis #AFISsys – an #autonomous instrument for the preservation of water samples for microbial #metatranscriptome analysis #HydroBios #EMBS55

The new issue of @genomeresearch is now live. Follow the link to new research on m6A RNA methylation following #SARSCoV2 infection, spatial #metatranscriptome and strand-specific RNA sequencing, and more! ow.ly/mKv050Nzeu8

To better comprehend #metagenome and #metatranscriptome sequencing applications, our technical specialist walked you through some fundamental concepts and case studies. Check out this video to learn more about the microbiome's role in health and disease! bit.ly/39Jjc2U

📢New Review alert for virus aficionados Current trends in RNA virus detection through metatranscriptome sequencing data 🔓buff.ly/3oy3u2e #RNAvirus #VirusDetection #Metatranscriptome #RNAvirome #Bioinformatics #Virology

Research published in @FrontMicrobiol investigates #metatranscriptome of eight social and solitary wild bee that reveals novel viruses and bee parasites: fron.tiers.in/go/GXRL8v

#SAMSA - a comprehensive #metatranscriptome analysis pipeline - bit.ly/2dDLZsq @genomecenter
Paper out in @BioMedCentral 1/ 🌱 We did a first #metatranscriptome study on #biofilm succession in the #soil #plastisphere and tracked #polyethylene (PE) plastics in soil over 53 days. Insights into biofilm dynamics, #plasticdegradation, and nitrogen fixation ahead! 🧵

#metatranscriptome の重要性①。静的( #Static )な遺伝子の状態をみるのではなく rRNA/mRNA 比を測定した上で、 動的な遺伝子状態 ( #metatranscriptome )を見なければ、内部の状態がどのようになっているかの予測がつかない。
#AFISsys - an autonomous instrument for the preservation of #brackishwater samples for microbial #metatranscriptome analysis sciencedirect.com/science/articl…

A 2024 study in Exp Ther Med used Magigene’s #Metatranscriptome sequencing service to analyze DSS-induced colitis, revealing XAV939 inhibits Wnt/β-catenin signal pathway. (Impact Factor: 2.4) 🔗Paper: spandidos-publications.com/10.3892/etm.20… 🔎Magigene’s solutions: en.magigene.com/wsw_1/13.html
See microbes in ACTION! 🔬 Magigene’s #Metatranscriptome skips culturing—analyze active RNA to: ✅ ID pathogens & pathways ✅ NovaSeq PE150 | 10G data 🌱🧪🌾 For soil/clinical/ag studies Explore → en.magigene.com/wsw_1/13.html #Microbiome #Biotech
Paper out in @BioMedCentral 1/ 🌱 We did a first #metatranscriptome study on #biofilm succession in the #soil #plastisphere and tracked #polyethylene (PE) plastics in soil over 53 days. Insights into biofilm dynamics, #plasticdegradation, and nitrogen fixation ahead! 🧵

Evaluation of commercial RNA extraction kits for long-read metatranscriptomics in soil. #nanopore #metatranscriptome doi.org/10.1099/mgen.0…

📢New Review alert for virus aficionados Current trends in RNA virus detection through metatranscriptome sequencing data 🔓buff.ly/3oy3u2e #RNAvirus #VirusDetection #Metatranscriptome #RNAvirome #Bioinformatics #Virology

The new issue of @genomeresearch is now live. Follow the link to new research on m6A RNA methylation following #SARSCoV2 infection, spatial #metatranscriptome and strand-specific RNA sequencing, and more! ow.ly/mKv050Nzeu8

In @CellCellPress: "#Metatranscriptome mining reveals a major expansion of #RNAVirus diversity" #microbiota #bacteriophage #virus #ViralEcology @UriNeri2 @ugophna @simroux_virus @mkrupovic @apcamargo_ @kyrpides @TelAvivUni @jgi @NCBI 👇 sciencedirect.com/science/articl…
Open #Bioinformatics Research Associate position (m/f/d) in the field of #metatranscriptome analysis in #leukemia @jlugiessen @FB08_JLU. For more information see: denbi.de/de-nbi-jobs-po…
Julian von Borries presenting an intriguing new gadget for your metatranscriptome analysis #AFISsys – an #autonomous instrument for the preservation of water samples for microbial #metatranscriptome analysis #HydroBios #EMBS55

To better comprehend #metagenome and #metatranscriptome sequencing applications, our technical specialist walked you through some fundamental concepts and case studies. Check out this video to learn more about the microbiome's role in health and disease! bit.ly/39Jjc2U

I am excited to share our recent research indicating that COVID-19 mortality is driven by two distinct trajectories # COVID-19 #Cytokine Release Syndrome-rare signature #metatranscriptome #Lung tissue journals.biologists.com/dmm/article/do…
Glad to share our last gut #microbiome study in collab with Chays Manichanh @vallhebron ⬇️ "Human gut #metatranscriptome changes induced by a fermented milk product are associated with improved tolerance to a flatulogenic diet" @DanoneResearch sciencedirect.com/science/articl…
Once we aligned the samples against the ref #metatranscriptome and normalized, based on a MDS plot it was clear that there was a strong signal across the first dimension. This indicated a strong effect of season, where early (June-July) and late (Aug-Oct) clustered separately...

Then once we chose the best reference (16 libs assembled together), we used this #metatranscriptome for all downstream applications, including aligning individual read files from each RNA-seq sample, as well as annotating for either putative function (SWISS-PROT) or taxonomy (nt)
A new item for me here was to try a few ways to assemble a ref #metatranscriptome. - incr. library inclusion step-by-step (1, 2, 4, 8, 12, 16) - assembling 8 libraries individually, then merge ...then we eval. the effects on length, contiguity, % align, and redundant align

#NovogeneWebinar In this webinar, our product marketing manager from @Novogene_Europe will guide you through the basic principles and case studies to help you understand #metagenome and #metatranscriptome applications. Click here for more information: bit.ly/3dN7eUe

Most-read analytica news: Listening in on laundry germs as they grow 🧺🦠 First #metatranscriptome study for washed #laundry bit.ly/3nq27j2 @hs_furtwangen @HenkelPresse @jlugiessen #analysis #RNASequencing #LaundryHygiene #textiles #clothing 📸 Furtwangen University

this is a very good point, thank you @DrKathrynElmer. I think I will go with that. Raw data is going on @NCBI's SRA, then the #metatranscriptome assembly on @datadryad.
Congrats to our postdoc fellow @YanYan20444341 for awarding the Charles King Trust Fellowship! Yan will study the strain-level epidemiology and diet-linked gut #metatranscriptome risk factors in #colorectal cancer.

just re-calculated the depth of our published #metatranscriptome Covering transcripts from species at <1% abundance!

@LauraTreu presenting the #genome-centric #metatranscriptome study on #Anaerobic #digestion system at #SIMGBM in Florence 2019! Great work! @DiBio_UniPD @IriniAngelidaki @kougias


#BIO2015 World Fusion's Tokyo division busy in booth 3649 with LSKB and drag&drop #metagenome & #metatranscriptome

Best place to upload a #metatranscriptome #assembly? (i.e., #RNAseq reads from environmental sample, assembled into a multiple-species assembly?) I figure this is useful for reproducibility, but possibly @NCBI is not the location? #eRNA ncbi.nlm.nih.gov/genbank/tsa/ Any ideas? Thanks!

The new issue of @genomeresearch is now live. Follow the link to new research on m6A RNA methylation following #SARSCoV2 infection, spatial #metatranscriptome and strand-specific RNA sequencing, and more! ow.ly/mKv050Nzeu8

📢New Review alert for virus aficionados Current trends in RNA virus detection through metatranscriptome sequencing data 🔓buff.ly/3oy3u2e #RNAvirus #VirusDetection #Metatranscriptome #RNAvirome #Bioinformatics #Virology

Julian von Borries presenting an intriguing new gadget for your metatranscriptome analysis #AFISsys – an #autonomous instrument for the preservation of water samples for microbial #metatranscriptome analysis #HydroBios #EMBS55

Once we aligned the samples against the ref #metatranscriptome and normalized, based on a MDS plot it was clear that there was a strong signal across the first dimension. This indicated a strong effect of season, where early (June-July) and late (Aug-Oct) clustered separately...

A new item for me here was to try a few ways to assemble a ref #metatranscriptome. - incr. library inclusion step-by-step (1, 2, 4, 8, 12, 16) - assembling 8 libraries individually, then merge ...then we eval. the effects on length, contiguity, % align, and redundant align

Research published in @FrontMicrobiol investigates #metatranscriptome of eight social and solitary wild bee that reveals novel viruses and bee parasites: fron.tiers.in/go/GXRL8v

#metatranscriptome analysis of DsrABC-sulfite reduction and DHPS/isethionate sulfite-lyase activities suggested Bilophila and other Desulfovibrionaceae use mostly #taurine but also #DHPS for #H2S production in the human gut

Evaluation of commercial RNA extraction kits for long-read metatranscriptomics in soil. #nanopore #metatranscriptome doi.org/10.1099/mgen.0…

Offre de #These #Etude #Metatranscriptome de la digestion #Anaerobie pour @Veolia_FR bit.ly/2wnS2Ik Durée 36 mois à pourvoir #ASAP

To better comprehend #metagenome and #metatranscriptome sequencing applications, our technical specialist walked you through some fundamental concepts and case studies. Check out this video to learn more about the microbiome's role in health and disease! bit.ly/39Jjc2U

#NovogeneWebinar In this webinar, our product marketing manager from @Novogene_Europe will guide you through the basic principles and case studies to help you understand #metagenome and #metatranscriptome applications. Click here for more information: bit.ly/3dN7eUe

Paper out in @BioMedCentral 1/ 🌱 We did a first #metatranscriptome study on #biofilm succession in the #soil #plastisphere and tracked #polyethylene (PE) plastics in soil over 53 days. Insights into biofilm dynamics, #plasticdegradation, and nitrogen fixation ahead! 🧵

Most-read analytica news: Listening in on laundry germs as they grow 🧺🦠 First #metatranscriptome study for washed #laundry bit.ly/3nq27j2 @hs_furtwangen @HenkelPresse @jlugiessen #analysis #RNASequencing #LaundryHygiene #textiles #clothing 📸 Furtwangen University

#AFISsys - an autonomous instrument for the preservation of #brackishwater samples for microbial #metatranscriptome analysis sciencedirect.com/science/articl…

Something went wrong.
Something went wrong.
United States Trends
- 1. Falcons 12.6K posts
- 2. Drake London 2,020 posts
- 3. Max B 11.9K posts
- 4. Raheem Morris N/A
- 5. Kyle Pitts 1,181 posts
- 6. Alec Pierce 2,016 posts
- 7. #Colts 2,494 posts
- 8. Penix 2,466 posts
- 9. Bijan 2,319 posts
- 10. $SENS $0.70 Senseonics CGM N/A
- 11. Badgley N/A
- 12. $LMT $450.50 Lockheed F-35 N/A
- 13. #ForTheShoe 1,408 posts
- 14. Zac Robinson N/A
- 15. $APDN $0.20 Applied DNA N/A
- 16. #Talus_Labs N/A
- 17. Good Sunday 75.6K posts
- 18. #DirtyBirds N/A
- 19. #AskFFT N/A
- 20. Jessie Bates N/A