#recombinasepolymeraseamplification результаты поиска
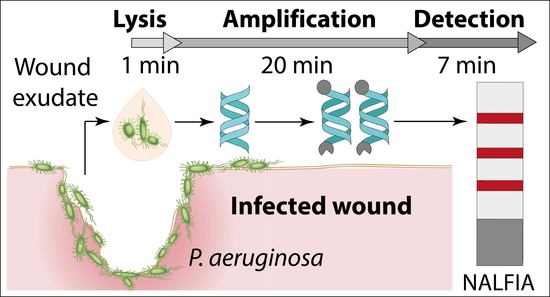
Rapid Detection of #Pathogens in Wound Exudate via Nucleic Acid Lateral Flow #Immunoassay Available at: mdpi.com/2079-6374/11/3… #recombinasepolymeraseamplification #pointofcarediagnostics #paperbaseddetection
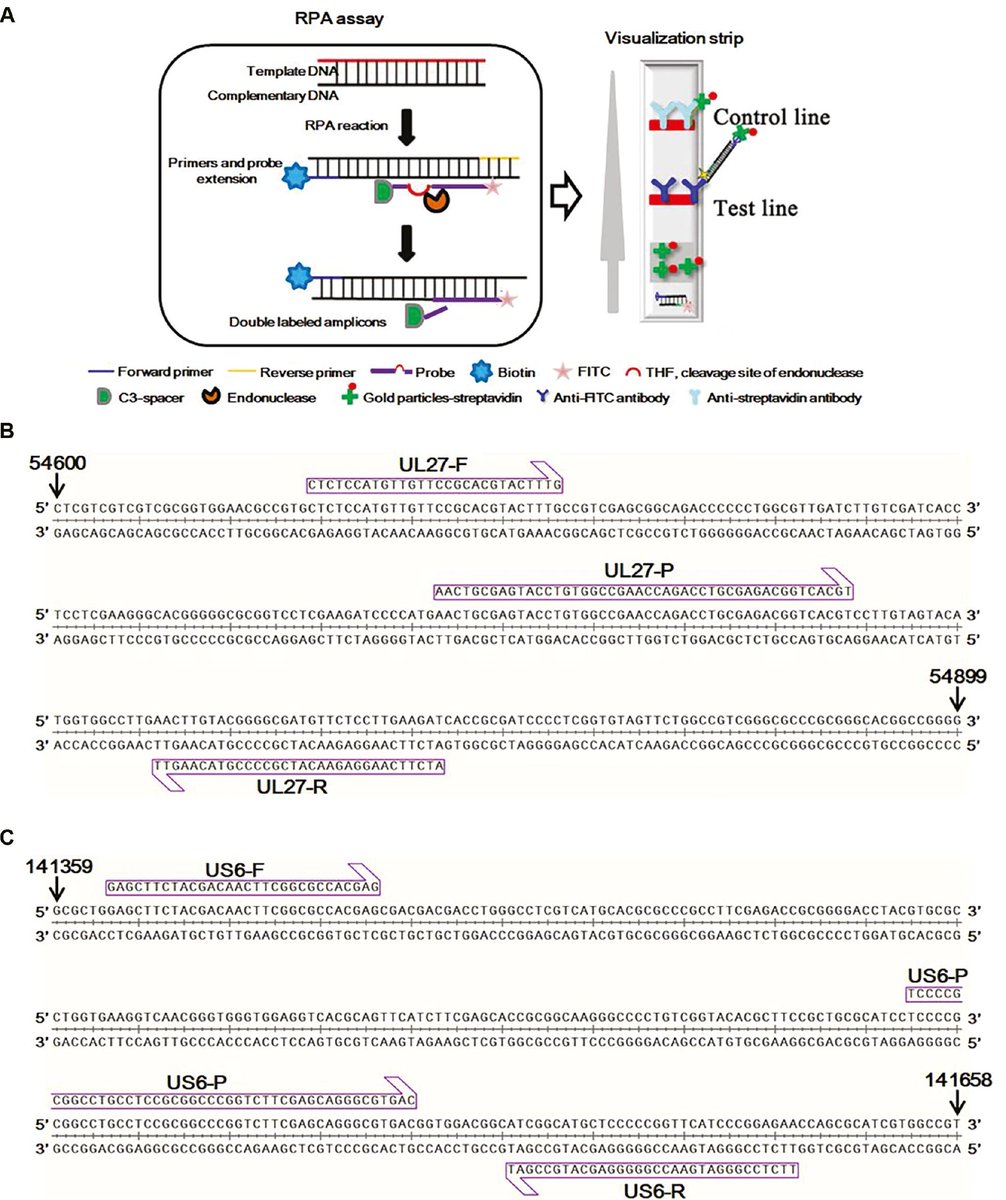
Rapid and Visual Detection of Monkey B Virus Based on Recombinase Polymerase Amplification #bvdiagnosis #recombinasepolymeraseamplification #poctesting zoonoses-journal.org/index.php/2023…
Today in @NatureBiotech, we report engineered DNA recombinases with up to 53% integration efficiency and 97% genome-wide specificity at an endogenous human locus, inserting large DNA cargoes up to 12 kb for stable expression in primary human T cells, stem cells, and non-dividing…
Site-specific DNA insertion into the human genome with engineered recombinases go.nature.com/43kdvBg

Genomes encode biological complexity, which is determined by combinations of DNA mutations across millions of bases In new @arcinstitute work, we report the discovery and engineering of the first programmable DNA recombinases capable of megabase-scale human genome rearrangement
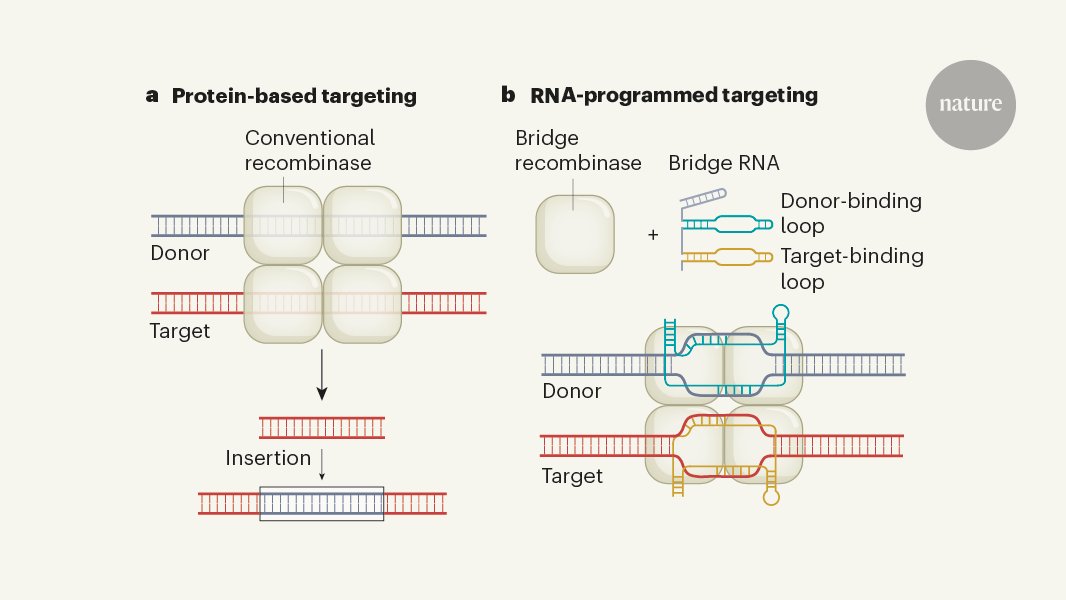
What if we could universally recombine, insert, delete, or invert any two pieces of DNA? In back-to-back @Nature papers, we report the discovery of bridge RNAs and 3 atomic structures of the first natural RNA-guided recombinase - a new mechanism for programmable genome design
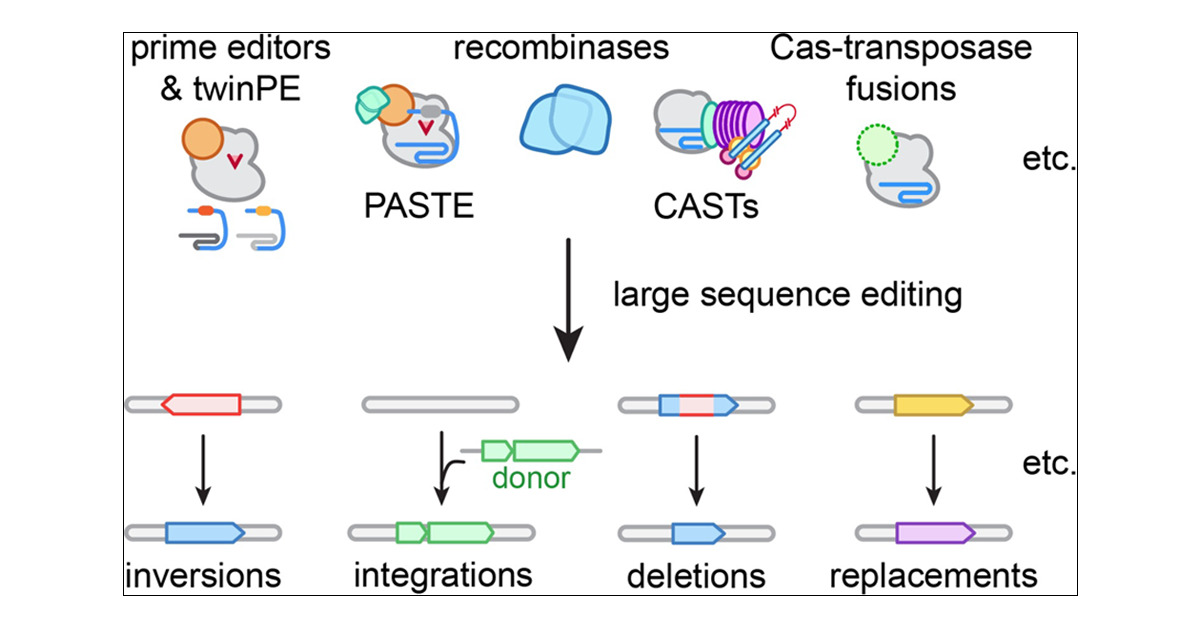
In the latest issue! Amplification editing enables efficient and precise duplication of DNA from short sequence to megabase and chromosomal scale dlvr.it/TBBMjh
A new chapter for genome editing: RNA-guided recombinase enzymes have been discovered that enable the programmable insertion, inversion or deletion of long DNA sequences go.nature.com/3RHgWMA
🧬 𝙋𝙤𝙡𝙮𝙢𝙚𝙧𝙖𝙨𝙚 𝘾𝙝𝙖𝙞𝙣 𝙍𝙚𝙖𝙘𝙩𝙞𝙤𝙣 🧬 "D.A.E. 957" Denaturation – 𝟗5°C Annealing – 𝟓2°C–55°C Extension – 𝟕2°C Process is repeated: 30 𝙩𝙞𝙢𝙚𝙨

What if you could recombine any two DNA sequences in a programmable manner? We discovered an RNA-guided recombinase that can do just that! So happy to have co-led this project with @ntperry13
Just shared at @KeystoneSymp a new @ArcInstitute discovery of the bridge RNA recombinase mechanism: a new class of natural RNA-guided systems that retains the key property of programmability from RNAi and CRISPR while enabling large-scale genome design beyond RNA and DNA cuts

Reverse transcriptase and recombinant DNA is what will continue the C19 synthetic genetic codes down through your legacy, for generations. Dr. Daniel Nagase explains it well.
Rapid and Visual Detection of Monkey B Virus Based on Recombinase Polymerase Amplification #bvdiagnosis #recombinasepolymeraseamplification #poctesting zoonoses-journal.org/index.php/2023…

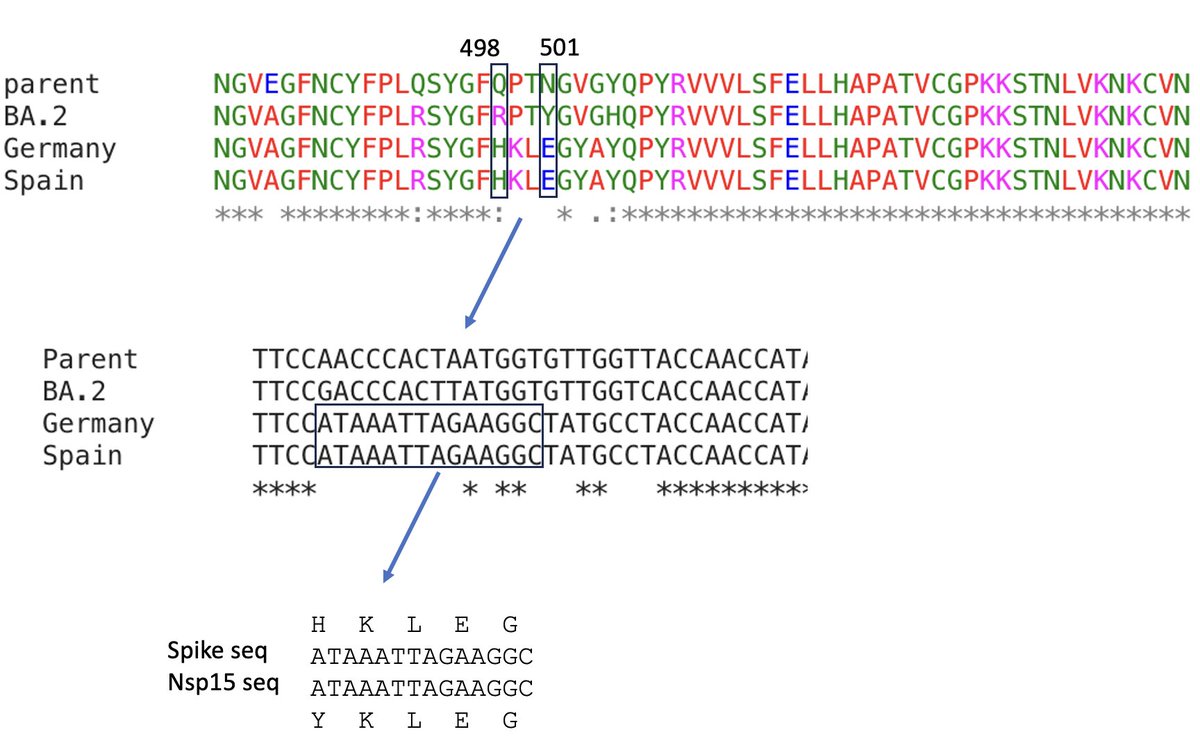
I showed this to @LongDesertTrain and he was a bit skeptical. He noted that the sequence was questionable with some of the sequences. Then he noticed something important. The sequence for a 5 amino acid portion of the RBD was a duplication of sequence from nsp15 in one seq. 6/
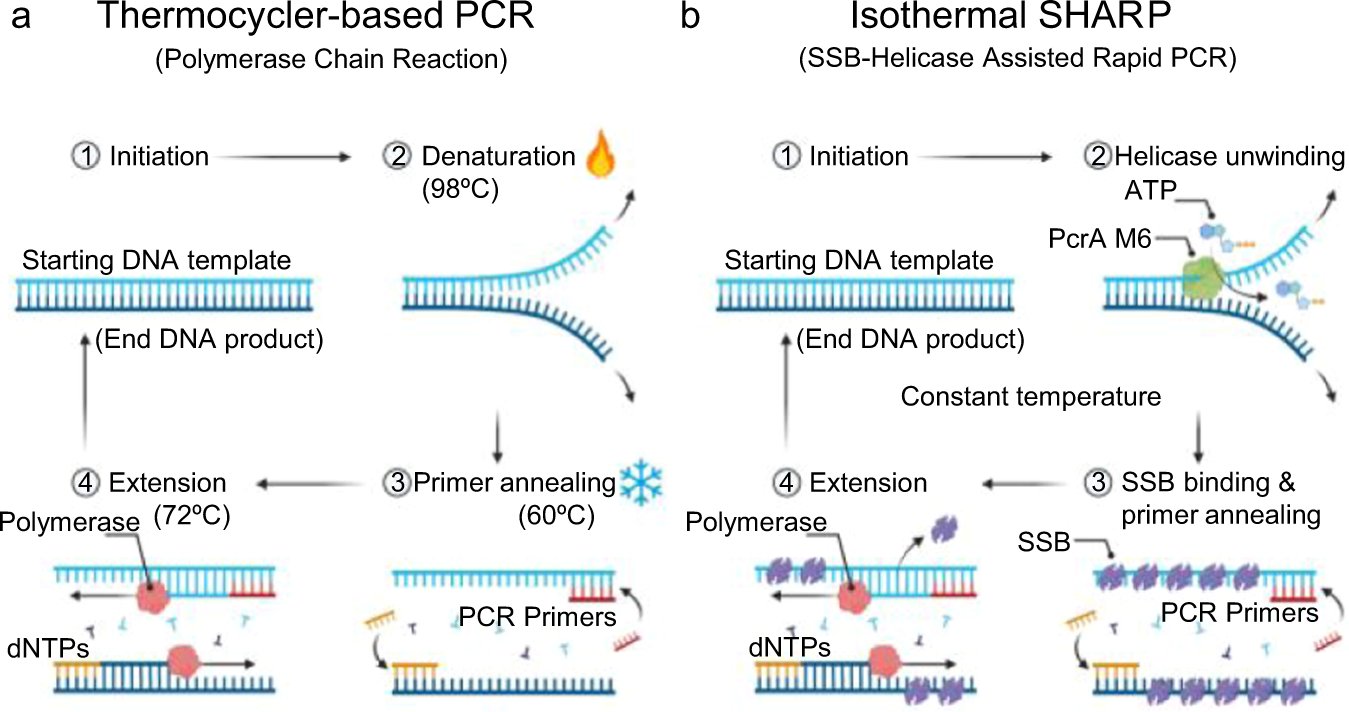
Isothermal DNA amplification up to 6 kb, led by @MomciloGavrilov ; can be used for cloning and chromosomal gene editing readout. A 3x faster superhelicase, characterized using nanopore and MD simulation (by Wen Ma), replaces thermocycler. nature.com/articles/s4146…
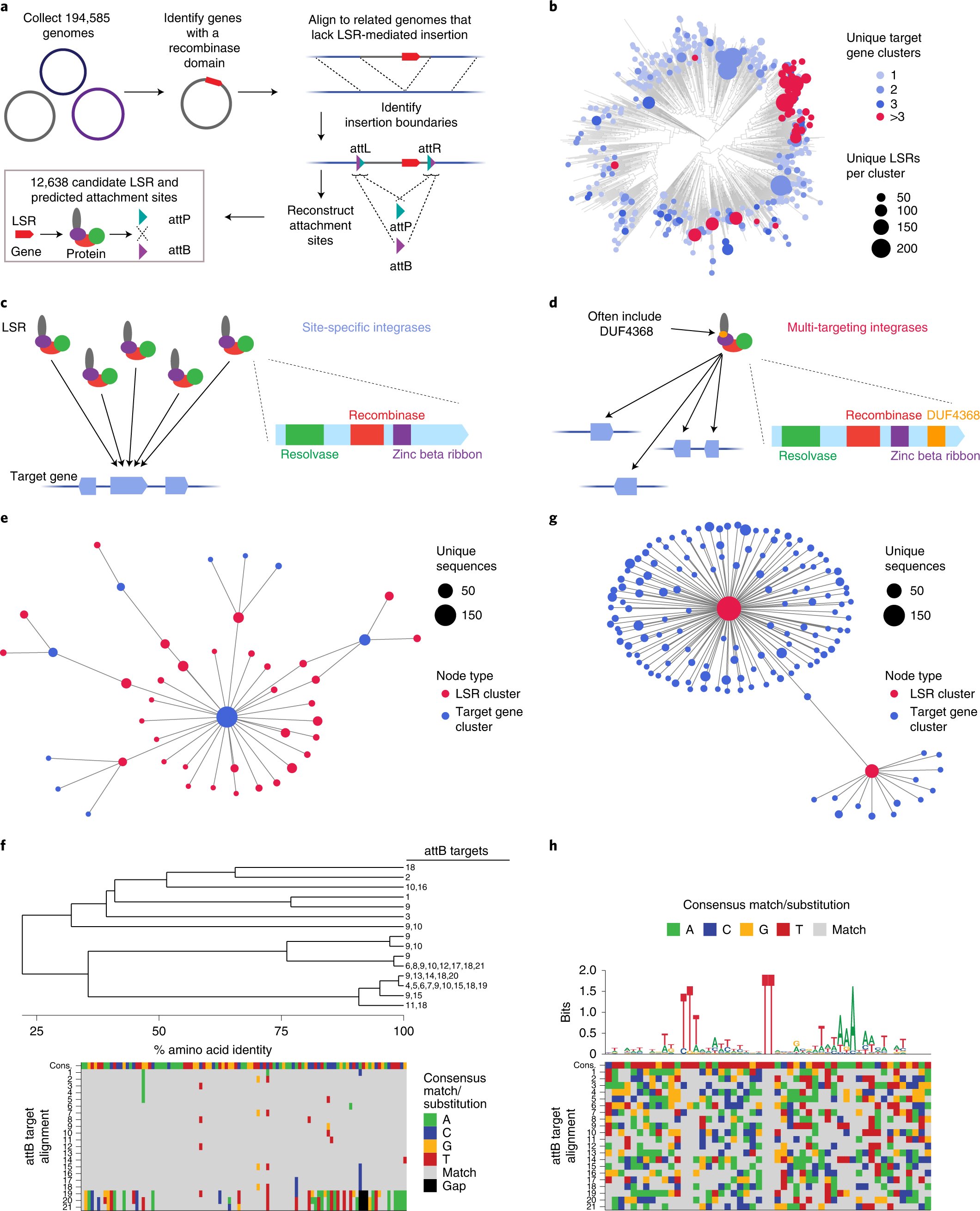
Our paper is out! New recombinases for large DNA integration in human cells nature.com/articles/s4158… We discovered thousands of recombinases and their DNA targets by searching in mobile genetic elements🧬🦠 Our experiments showed some are promising tools for genome engineering🖋️
Today in @BiochemistryACS, we share a perspective on emergent kilobase-scale genome editing technologies - including dual-flap prime editing, recombinases, and transposases (+ their variations). pubs.acs.org/doi/10.1021/ac…
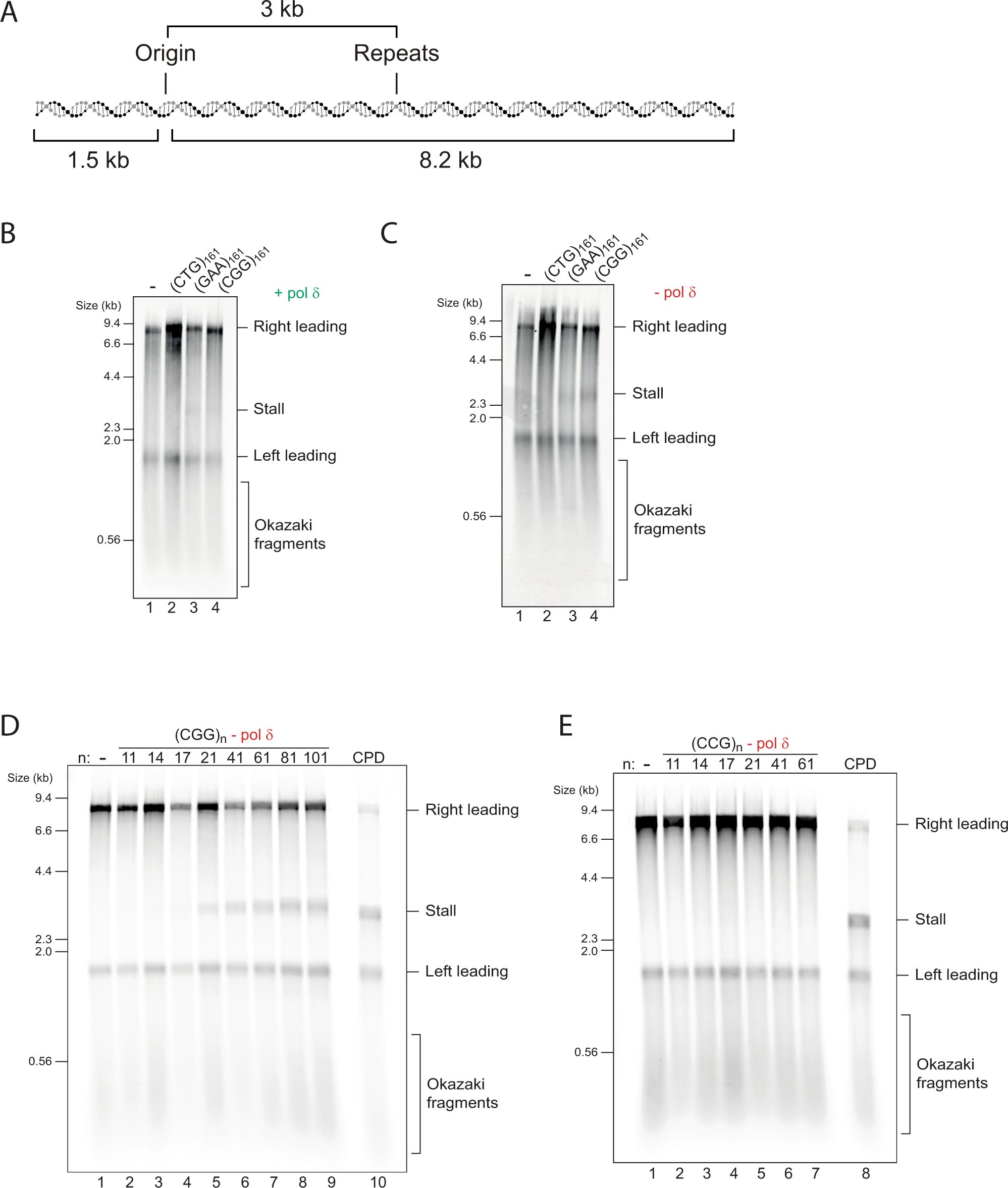
📢 Hot off the press 📢 🧬 Ever wonder why some DNA sequences are difficult to replicate? Using reconstituted replisomes we revealed that structure-forming repeats induce robust replication stalling and uncovered structure-specific recovery mechanisms: nature.com/articles/s4146…
Die RNA kann man aber in der PCR nicht verdoppeln, weil man damit nicht exponentiell vermehren kann. Dafür braucht man 2 Stränge. Und dafür muss man die RNA in DNA umschreiben. Und dafür wiederum braucht man die Reverse Transkriptase. Ein Enzym.
A new method developed to reread a single polypeptide many times overcomes one of the biggest obstacles toward single-molecule proteomics. Read more in a new #SciencePerspective: fcld.ly/23d7ov4
Rapid Detection of #Pathogens in Wound Exudate via Nucleic Acid Lateral Flow #Immunoassay Available at: mdpi.com/2079-6374/11/3… #recombinasepolymeraseamplification #pointofcarediagnostics #paperbaseddetection

A new method developed to reread a single polypeptide many times overcomes one of the biggest obstacles toward single-molecule proteomics. Read more in a new #SciencePerspective: fcld.ly/23d7ov4
Except there is an extra step. The COVID genome is not made of DNA, it’s actually RNA, which only comes in single strands. So we add another enzyme, called Reverse Transcriptase, which replaces the missing strand and turns it into double stranded DNA for PCR. 9/n
Cool it down again, and the enzyme polymerase (the P in PCR) fills in the rest, so you end up with two identical, photocopied versions of the original bit of DNA. GAACTTAATTAA CTTGAATTAATT GAACTTAATTAA CTTGAATTAATT 6/n
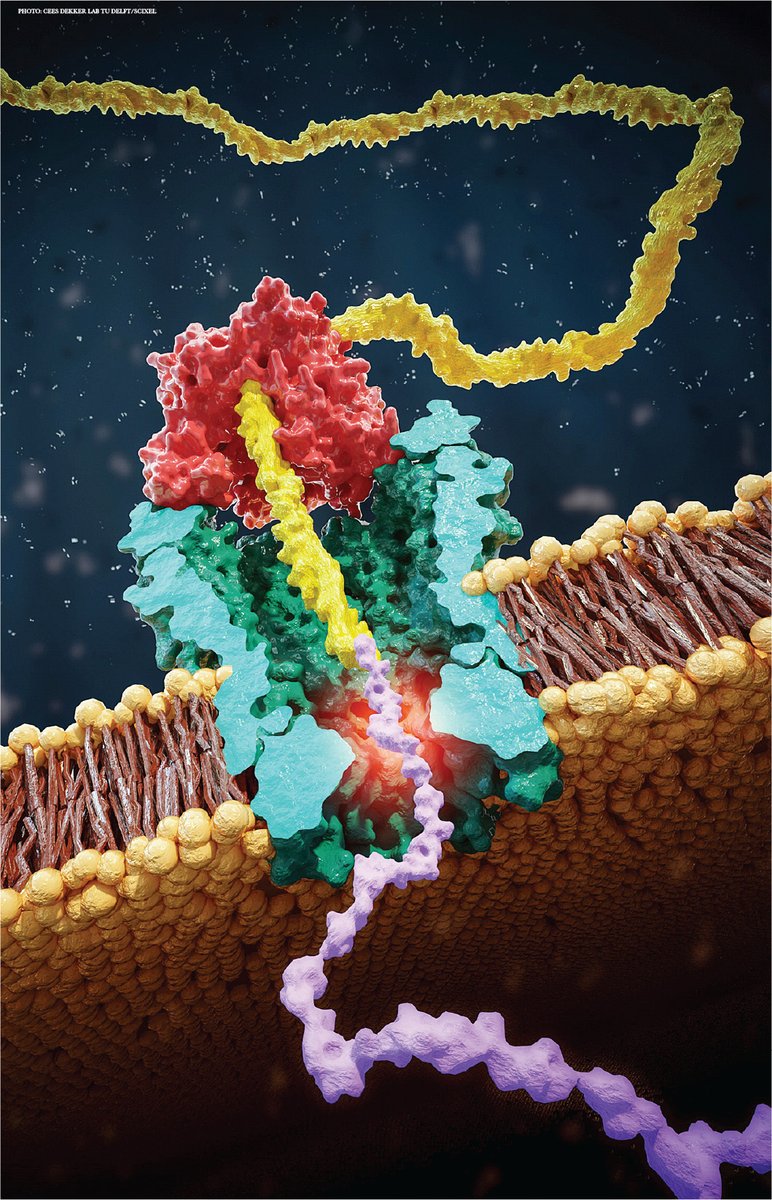
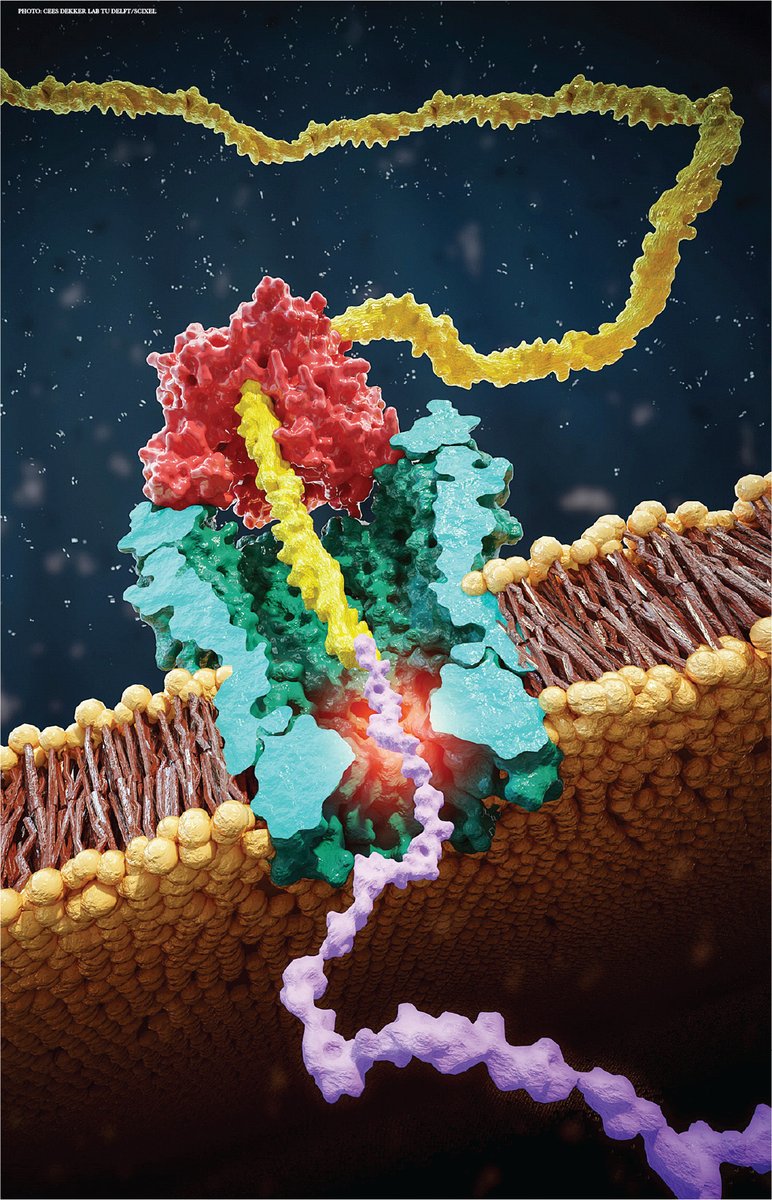
WOW: T cells take up (via extracellular vesicles) telomeres from antigen presenting cells at the immunological synapse. Telomere vesicles retained the Rad51 recombination factor enabling fusing with T cell chromosomal ends causing an average lengthening of ~3000 base pairs 🤯🤯
Intercellular telomere transfer extends T cell lifespan disq.us/t/3s5vtlp Why do T cells give up their specific receptors in vesicles? Alessio and team find that they are "traded" with APC for telomeres that increase T cell lifespan. Exited to be part of the team!
Rapid Detection of #Pathogens in Wound Exudate via Nucleic Acid Lateral Flow #Immunoassay Available at: mdpi.com/2079-6374/11/3… #recombinasepolymeraseamplification #pointofcarediagnostics #paperbaseddetection

Rapid and Visual Detection of Monkey B Virus Based on Recombinase Polymerase Amplification #bvdiagnosis #recombinasepolymeraseamplification #poctesting zoonoses-journal.org/index.php/2023…

Something went wrong.
Something went wrong.
United States Trends
- 1. #WWENXT 6,969 posts
- 2. Notre Dame 29.8K posts
- 3. Notre Dame 29.8K posts
- 4. Bama 20K posts
- 5. Jaylen Brown 2,816 posts
- 6. Van Epps 107K posts
- 7. Paul Dano 4,252 posts
- 8. Ament N/A
- 9. Cuse 1,348 posts
- 10. Cam Boozer N/A
- 11. Behn 62.1K posts
- 12. Thomas Haugh N/A
- 13. #CFPRankings N/A
- 14. Tarantino 11K posts
- 15. #TADCFriend 2,605 posts
- 16. Hugo Gonzalez N/A
- 17. Penn State 20.8K posts
- 18. Jaylen Carey N/A
- 19. Meechie Johnson N/A
- 20. Stirtz N/A