#spatialtranscriptomic résultats de recherche
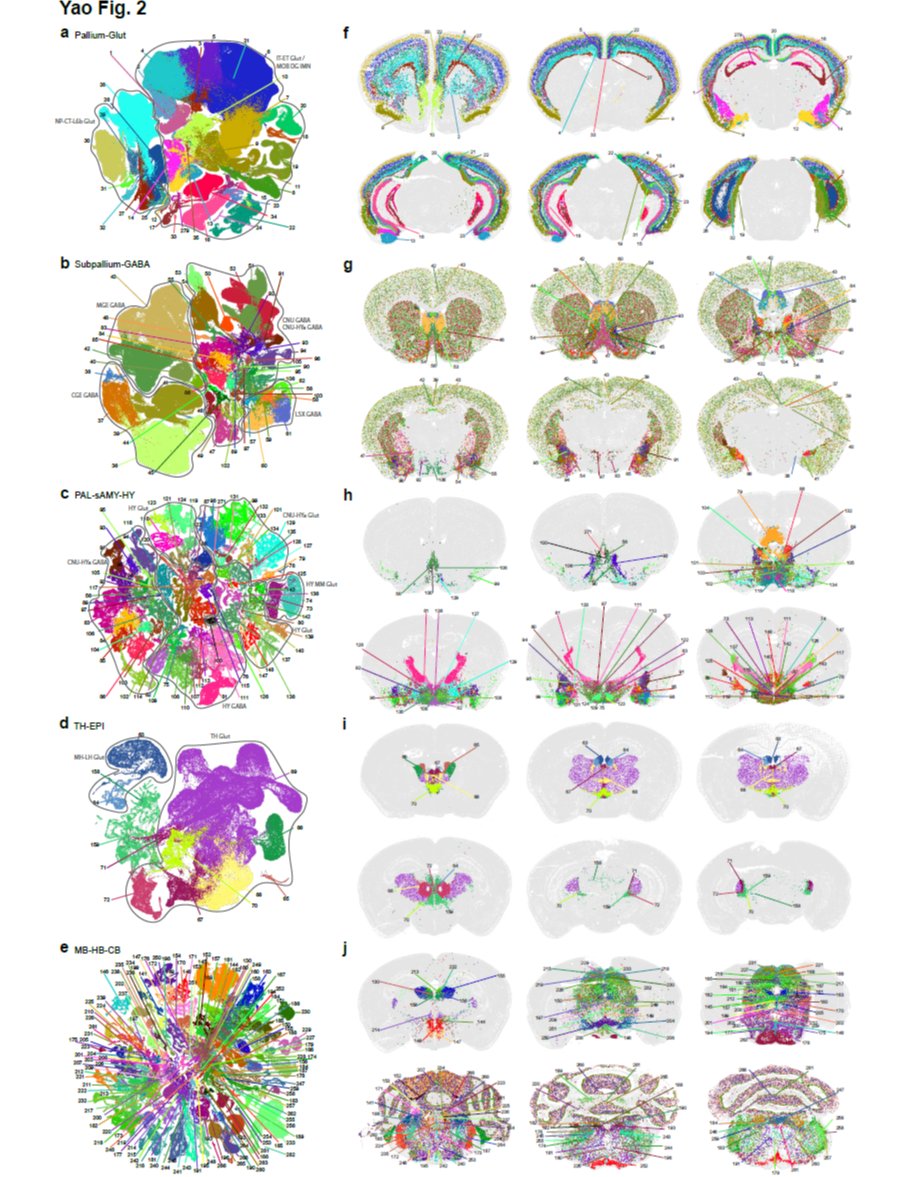
A🐭🧠 project spearheaded by @AllenInstitute created a #SpatialTranscriptomic #CellTypeAtlas of the whole #MouseBrain! Many key marker genes used in our new #MERSCOPE PanNeuro Cell Type Panel were validated in this project. Read the paper & learn more: hubs.ly/Q01KGWK90
@mason_lab talks about #spatialtranscriptomic profiling of tissues infected with #SARS #COVID19 #AGBT21

Thrilled to receive the NIH/NIGMS R35 MIRA to support our #bioinformatics work studying subcellular and cellular #spatialtranscriptomic heterogeneity Special thanks to @rajivmccoy for guidance + support from mentors @jtleek, Joel Bader, Mike Miller @JHUBME 🥳🥂 #AcademicChatter

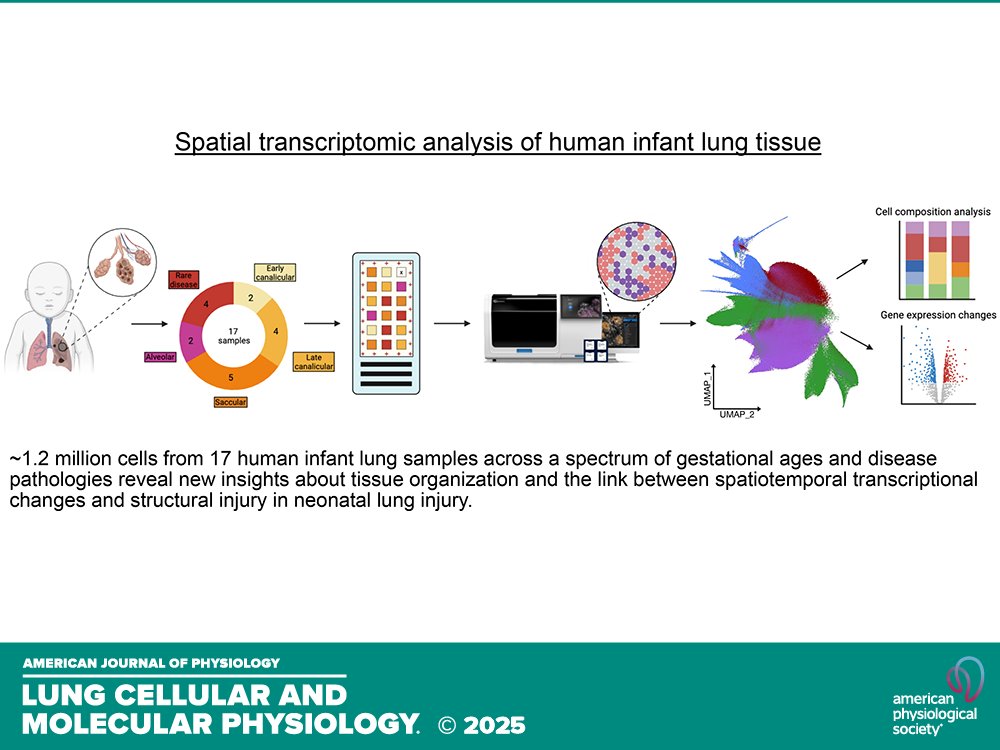
Short Report by S Mallapragada et al. (@AnnikaVannan @ASU @TGenResearch @VUMChealth @UW) A #spatialtranscriptomic atlas of acute #neonatal #lunginjury across development and disease severity ow.ly/nYfo50WMbH5
Starting today....@nanostringtech Nanostring GEOMx fully operational and training beginning! @UofGMVLS @pvanhouts Mathias Holpert @UofGCancerSci @CRUKGlasgow looking forward to exciting #spatialtranscriptomic projects @CRUK_BI @grisurgery @hollyleslie_ @EdinCRC @precisionpanc




1/4 #SpatialTranscriptomic methods allow to analyse the gene expression of tissues without losing its spatial heterogeneity. In my #PhD in the Meier lab @PioneerCampus I apply and advance such methods to address different biological questions. #YoungScientistsHMGU

@OliverStegle presenting on identification of spatially variable genes #spatialtranscriptomic #cytodata2019

Welcome to Tu Duyen Nguyen, @AllianceProg intern from @Polytechnique. Tu Duyen is joining the IICD to focus on novel algorithms to understand the spatial interaction between cells from #spatialtranscriptomic data in the @elhamazizi lab.

Check out 🔻🔻 #cholangiocarcinoma Paper from @chiarabraconi @ColmJJORourke gut.bmj.com/content/early/… Well done @LabSpatialNBJ for #spatialtranscriptomic component @Gut_BMJ @grisurgery @nanostringtech @UofGMVLS @UofGCancerSci


The first installation of @nanostringtech #cosmx in the UK 🇬🇧 explains all the big smiles! Looking forward to the incredible #singlecell #spatialtranscriptomic data that @LabSpatialNBJ team will produce @UofGCancerSci @UofGMVLS @UofGlasgow Thanks @The_MRC @CRUKresearch



Friday afternoon fun going through my first @nanostringtech #GeoMx images. Trying to optimise morphology markers before embarking on the first #SpatialTranscriptomic experiment to be run @cardiffuni

STMiner, an algorithm specifically designed for complex #spatialtranscriptomic data can remove false-positive genes from the analysis results. We are deeply appreciative of @CellGenomics for affording us the platform to present this significant research outcome.

Have you had the chance to explore what a #MERFISHmeasurement dataset from our #MERSCOPE in situ #SpatialTranscriptomic platform looks like? Take a break today and access the MERFISH Mouse Brain Receptor Map! hubs.ly/H0SWCFH0
Sharing our experience about spatial transcriptomic in thyroid lesions. @ThaisMaloberti #spatialtranscriptomic #thyroidtumors

Our lab student Kaan Cifcibasi presenting his highly interesting #spatialtranscriptomic data in PDAC @German_PancClub ! Meet us at the Congress! #pancreaticcancer #cancer #pankreasclub #bonn #demirlab

In December 2020, Dr. Zhuang's lab at @Harvard posted an article to @biorxivpreprint detailing how they used #MERFISH to generate a subcellular #SpatialTranscriptomic atlas of neurons. Read & learn about the patterns of #RNA in neurons. hubs.ly/H0-cgx90 #ThrowbackThursday

Last week has been the most exciting in our Spatial Genomic Team -at @sangerinstitute , for beta-testing the N-01 @10xGenomics #CytAssist with #Visium FFPE Version2 I’m so grateful and honoured to have actually run our samples in the first model!! 🔬👩🏻🔬❤️#spatialtranscriptomic




📷#MERSCOPE generates #MERFISH #SpatialTranscriptomic #DataImages that are not only exceptional in their #Accuracy & #DetectionEfficiency, but also breathtaking in their beauty.🎨 Check out the winners of our 2024 calendar contest: hubs.ly/Q02k1LM40 #VizgenViews #SciArt

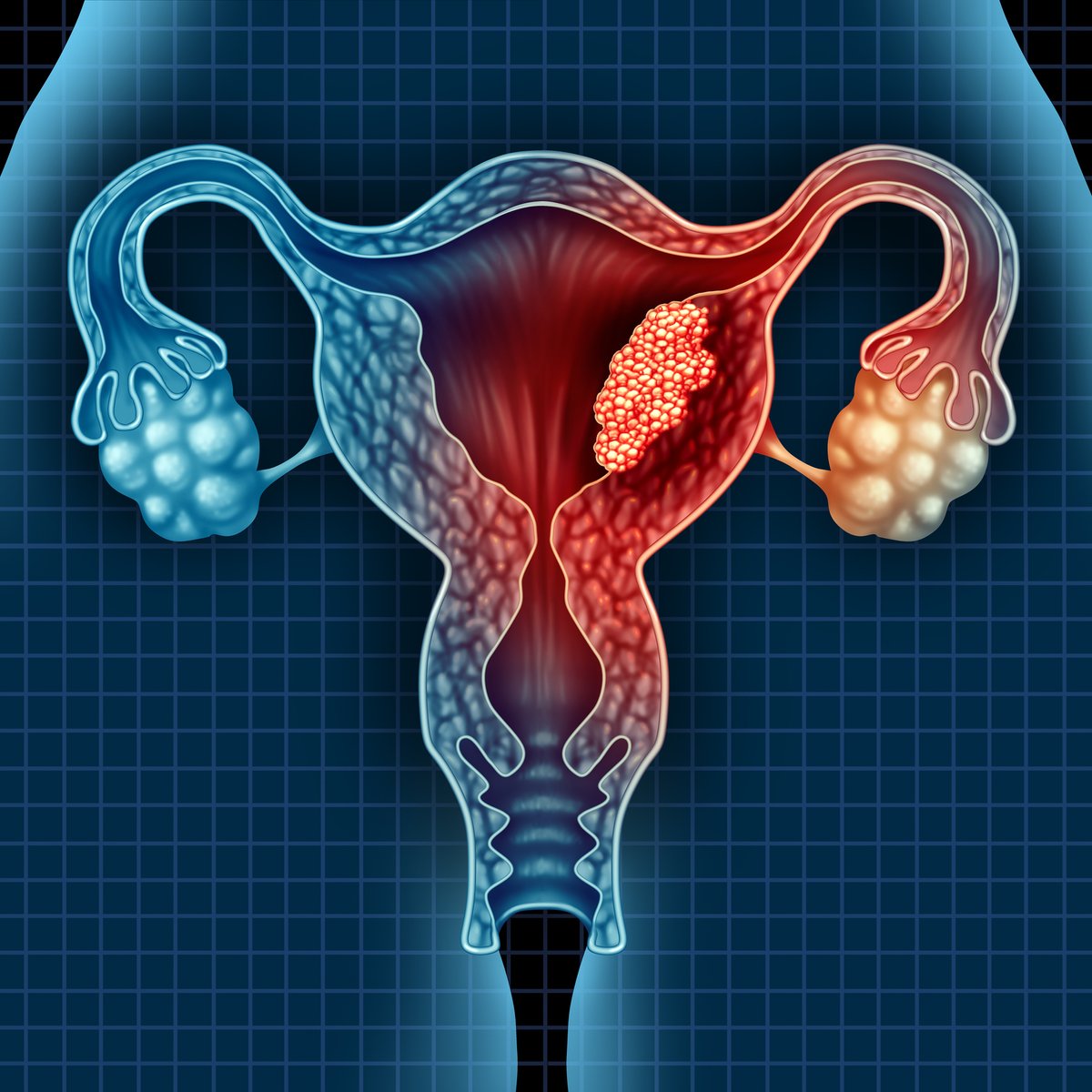
The first in-depth reference map of the uterus to collect both single cell and #spatialtranscriptomic data reveals new insights into the cellular make up of #endometrialcancers and adds to the Human Cell Atlas. Learn more: ow.ly/f5Xz50H35bQ #uterinecancer
Our summit in Paris is running successfully today; thanks to all our great speakers #spatialtranscriptomic

Advice needed: can you distinguish splicing variants by commercial #spatialtranscriptomic (eg visium)? #singlecell #omics
Short Report by S Mallapragada et al. (@AnnikaVannan @ASU @TGenResearch @VUMChealth @UW) A #spatialtranscriptomic atlas of acute #neonatal #lunginjury across development and disease severity ow.ly/nYfo50WMbH5

Post-doctoral researcher in oncology and computer science Paris Brain Institute See the full job description on jobRxiv: jobrxiv.org/job/paris-brai… #NeuroOncology #radiomic #spatialtranscriptomic #ScienceJobs jobrxiv.org/job/paris-brai…
Fascinating study from prof @ZeminZhangLab from Peking University fresh out in @Nature using both #scrna & #spatialtranscriptomic to decipher cross-tissue multicellular coordination and rewiring in cancer!
Nature research paper: Cross-tissue multicellular coordination and its rewiring in cancer go.nature.com/3ZEdXYZ
Excited to share my latest @JHepatology editorial on this impactful @Nature paper by my good friends @JoeYeong & Prof Liu Lianxin—highlighting Singapore–China collaboration & the power of #spatialtranscriptomic in predicting #HCC #recurrence. sciencedirect.com/science/articl…
Nice talk from Tarine Standgaard on tumour heterogeneity in #NMIBC & use of #SpatialTranscriptomic (& bulk) analyses to distinguish BCG-responsive molecular subtype So gd to finally see #EAUlab up and running!! Thanks #EAU25 committee & #ESUR @Uroweb @LauraBukavinaMD @LDyrskjot



STMiner, an algorithm specifically designed for complex #spatialtranscriptomic data can remove false-positive genes from the analysis results. We are deeply appreciative of @CellGenomics for affording us the platform to present this significant research outcome.

Caractéristiques spatiales et sensibilité aux traitements de 10 indications tumorales. #spatialtranscriptomic. The spatial landscape of cancer hallmarks reveals patterns of tumor ecological dynamics and drug sensitivity: Cell Reports cell.com/cell-reports/f…
The latest technical blog from scientists at the Earlham Institute shares some insights, top tips, and a workflow for applying #spatialtranscriptomic technologies to plant tissue. 🔎🌿🧬 ➡️ okt.to/ujacFs #PlantScience

Our CYGNUS project is developing a powerful new pipeline for the detailed study of diagnostic and archival tissue samples, enabling researchers to explore #SpatialTranscriptomic data 🔬 Discover more at buff.ly/4eg0bky Image by Jamieson Spatial Lab

🧬 Scientists at the Earlham Institute are developing advanced #singlecell and #spatialtranscriptomic approaches to explore gene expression in plant and animal systems, uncovering the mechanisms they use to adapt to challenges. #CellularGenomics okt.to/e73nFf
earlham.ac.uk
Cell expression heterogeneity impact on environmental response
Linking heterogeneity in gene expression within individual cells with whole organism phenotypes and adaptability.
Neuronal heterogeneity in the medial septum and diagonal band of Broca: classes and continua biorxiv.org/content/10.110… #MERFISH #spatialtranscriptomic #singlecell #RNA #MERSCOPE @vizgen_inc
Discover imaging solutions at @MedDiscCat, transforming drug discovery and enhancing therapeutic effectiveness. 💊 From #MassSpectrometryImaging to #SpatialTranscriptomic capability, unlock the potential of integrated data to elevate development. Read👉 loom.ly/utgKgk4

Sharing our experience about spatial transcriptomic in thyroid lesions. @ThaisMaloberti #spatialtranscriptomic #thyroidtumors

Excited to share this fantastic opportunity for an intensive hands-on course in #spatialtranscriptomic with the @10xGenomics #Xenium @MRC_WIMM @UniofOxford. Perfect for those looking to expand their skills - don’t miss out! Unlock the secrets and register today 👇 #SingleCellOmic
Join us for a three-day hands-on course in Spatial Transcriptomics with the 10x Xenium at the @MRC_WIMM Advanced Single Cell Omics Facility (WASCOF) @UniofOxford. From Panel design & Tissue Preparation to Imaging & Data Analysis. Register your intetest: [email protected]


Our lab student Kaan Cifcibasi presenting his highly interesting #spatialtranscriptomic data in PDAC @German_PancClub ! Meet us at the Congress! #pancreaticcancer #cancer #pankreasclub #bonn #demirlab

Super excited to be a part of this amazing team, and looking forward to discovering more with the cutting edge #spatialtranscriptomic for lung cancer patients.
Grateful for the support from the MRFF and incredible researchers supporting this Lung Cancer #MetaSpatial study #teamscience @chinwee10 @Dr_MarkAdams @bhuva_dd @lnly0311 @ProfGabBelz @CCRG_Research @Qld_SBC @WesleyResearch @WEHI_research @TRI_info @SAiGENCI @UQ_News
📷#MERSCOPE generates #MERFISH #SpatialTranscriptomic #DataImages that are not only exceptional in their #Accuracy & #DetectionEfficiency, but also breathtaking in their beauty.🎨 Check out the winners of our 2024 calendar contest: hubs.ly/Q02k1LM40 #VizgenViews #SciArt

🎇For the first time ever, we propose Infinite #SpatialTranscriptomic editing using #GenerativeAI. Our approach enables algorithmic gene expression-guided editing in a generated gigapixel mouse pup🐭. Code, preprint and generated 106496 x 53248 WSIs: github.com/CTPLab/IST-edi…

Join us in Seattle on 10/26 at the @10xGenomics User Group Meeting for presentations from leading scientists & networking! Visit our table for a demo of Partek Flow and see how to easily analyze your #singlecell and #spatialtranscriptomic data. partek.com/10x_10.26.23_S…



A🐭🧠 project spearheaded by @AllenInstitute created a #SpatialTranscriptomic #CellTypeAtlas of the whole #MouseBrain! Many key marker genes used in our new #MERSCOPE PanNeuro Cell Type Panel were validated in this project. Read the paper & learn more: hubs.ly/Q01KGWK90

Thrilled to receive the NIH/NIGMS R35 MIRA to support our #bioinformatics work studying subcellular and cellular #spatialtranscriptomic heterogeneity Special thanks to @rajivmccoy for guidance + support from mentors @jtleek, Joel Bader, Mike Miller @JHUBME 🥳🥂 #AcademicChatter

@mason_lab talks about #spatialtranscriptomic profiling of tissues infected with #SARS #COVID19 #AGBT21

Check out 🔻🔻 #cholangiocarcinoma Paper from @chiarabraconi @ColmJJORourke gut.bmj.com/content/early/… Well done @LabSpatialNBJ for #spatialtranscriptomic component @Gut_BMJ @grisurgery @nanostringtech @UofGMVLS @UofGCancerSci


In December 2020, Dr. Zhuang's lab at @Harvard posted an article to @biorxivpreprint detailing how they used #MERFISH to generate a subcellular #SpatialTranscriptomic atlas of neurons. Read & learn about the patterns of #RNA in neurons. hubs.ly/H0-cgx90 #ThrowbackThursday

Starting today....@nanostringtech Nanostring GEOMx fully operational and training beginning! @UofGMVLS @pvanhouts Mathias Holpert @UofGCancerSci @CRUKGlasgow looking forward to exciting #spatialtranscriptomic projects @CRUK_BI @grisurgery @hollyleslie_ @EdinCRC @precisionpanc




1/4 #SpatialTranscriptomic methods allow to analyse the gene expression of tissues without losing its spatial heterogeneity. In my #PhD in the Meier lab @PioneerCampus I apply and advance such methods to address different biological questions. #YoungScientistsHMGU

Welcome to Tu Duyen Nguyen, @AllianceProg intern from @Polytechnique. Tu Duyen is joining the IICD to focus on novel algorithms to understand the spatial interaction between cells from #spatialtranscriptomic data in the @elhamazizi lab.

📷#MERSCOPE generates #MERFISH #SpatialTranscriptomic #DataImages that are not only exceptional in their #Accuracy & #DetectionEfficiency, but also breathtaking in their beauty.🎨 Check out the winners of our 2024 calendar contest: hubs.ly/Q02k1LM40 #VizgenViews #SciArt

The first in-depth reference map of the uterus to collect both single cell and #spatialtranscriptomic data reveals new insights into the cellular make up of #endometrialcancers and adds to the Human Cell Atlas. Learn more: ow.ly/f5Xz50H35bQ #uterinecancer

Now open! Research Award for #SpatialBiology across the #WholeTranscriptome. Two awards available: Human & Mouse. #SpatialTranscriptomic. In partnership with @EACRnews and @illumina. Apply today! ow.ly/Bfig50EwdaV

Short Report by S Mallapragada et al. (@AnnikaVannan @ASU @TGenResearch @VUMChealth @UW) A #spatialtranscriptomic atlas of acute #neonatal #lunginjury across development and disease severity ow.ly/nYfo50WMbH5

The first installation of @nanostringtech #cosmx in the UK 🇬🇧 explains all the big smiles! Looking forward to the incredible #singlecell #spatialtranscriptomic data that @LabSpatialNBJ team will produce @UofGCancerSci @UofGMVLS @UofGlasgow Thanks @The_MRC @CRUKresearch



Nice talk from Tarine Standgaard on tumour heterogeneity in #NMIBC & use of #SpatialTranscriptomic (& bulk) analyses to distinguish BCG-responsive molecular subtype So gd to finally see #EAUlab up and running!! Thanks #EAU25 committee & #ESUR @Uroweb @LauraBukavinaMD @LDyrskjot



Rao's team at the @QIMRBerghofer used GeoMx DSP to study #spatialtranscriptomic signatures and viral load in lungs from #SARS-CoV-2 infected hamsters and revealed that nuclear ACE2 may be a therapeutic target independent of the variant. 👉go.nature.com/42ZfAiR

@biotechne announces the commercial release of the #RNAscope HiPlex V2 Assay for #scRNAseq validation and #spatialtranscriptomic profiling while retaining tissue morphology! #enhancingyourspatialgenomicsstory #RNAISH #HiPlex Read the Press Release here: bit.ly/3zGLFhf

Single Cell RNA-Seq and Spatial Data Merge Using Cell2location A team has developed a tool, сell2location, that can resolve fine-grained cell types in #spatialtranscriptomic data and create comprehensive cellular maps of diverse tissues. Learn more: ow.ly/9Zya50HuEaq

Last week has been the most exciting in our Spatial Genomic Team -at @sangerinstitute , for beta-testing the N-01 @10xGenomics #CytAssist with #Visium FFPE Version2 I’m so grateful and honoured to have actually run our samples in the first model!! 🔬👩🏻🔬❤️#spatialtranscriptomic




Our lab student Kaan Cifcibasi presenting his highly interesting #spatialtranscriptomic data in PDAC @German_PancClub ! Meet us at the Congress! #pancreaticcancer #cancer #pankreasclub #bonn #demirlab

@OliverStegle presenting on identification of spatially variable genes #spatialtranscriptomic #cytodata2019

Something went wrong.
Something went wrong.
United States Trends
- 1. Veterans Day 386K posts
- 2. Woody 13.6K posts
- 3. Tangle and Whisper 1,936 posts
- 4. #stateofplay 9,202 posts
- 5. Toy Story 5 18.8K posts
- 6. Nico 144K posts
- 7. Luka 85.1K posts
- 8. Gambit 43.7K posts
- 9. Travis Hunter 4,035 posts
- 10. Payne 12.5K posts
- 11. Tish 5,660 posts
- 12. SBMM 1,560 posts
- 13. Square Enix 5,383 posts
- 14. Wike 126K posts
- 15. Mavs 33.3K posts
- 16. Sabonis 3,925 posts
- 17. De Minaur 3,261 posts
- 18. Vets 34.4K posts
- 19. Antifa 194K posts
- 20. Jonatan Palacios 2,383 posts




































