#structureprediction ผลการค้นหา
We are delighted to announce the release of structural models for 6,370 families of unknown structure provided by Baker group @UWproteindesign: xfam.wordpress.com/2021/03/03/fol… (funded by @BBSRC) #structureprediction #trRosetta #structuralbiology #deeplearning #AI

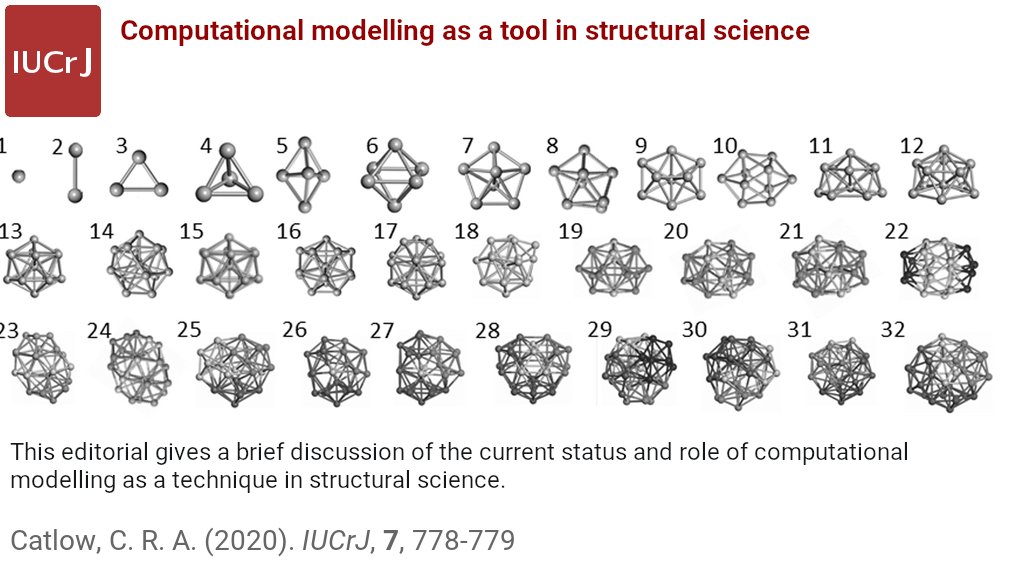
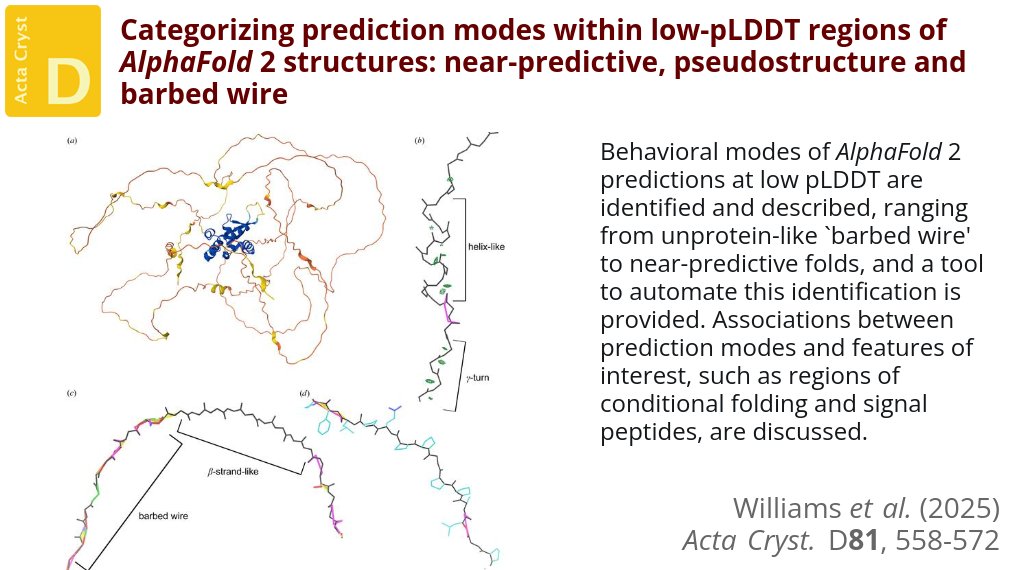
Major modes of behavior within low-pLDDT regions were identified through a survey of human proteome predictions provided by the AlphaFold Protein Structure Database @ActaCrystD @IUCr #AlphaFold2 #StructurePrediction #ConditionalFolding doi.org/10.1107/S20597…
Ok, so I decided to superimpose myosin structure to its alphafold predicted structure.. that went well.. #alphafold2 #Biochem #structureprediction
Advances in #RNA 3D Structure Prediction #RNAstructure #StructurePrediction #compchem pubs.acs.org/doi/10.1021/ac… #vol62 #issue23 #JCIM #Reviews
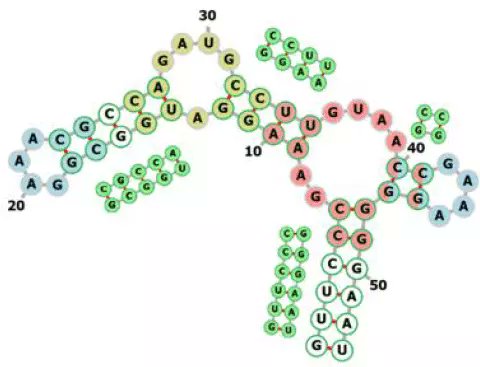
Scientists unveil #ProteinBench—a new evaluation framework to improve transparency in protein foundation models. With its multi-metric approach, it aims to drive innovation & collaboration in 3D #structureprediction, #proteindesign, and more! Quick Read: cbirt.net/proteinbench-u…
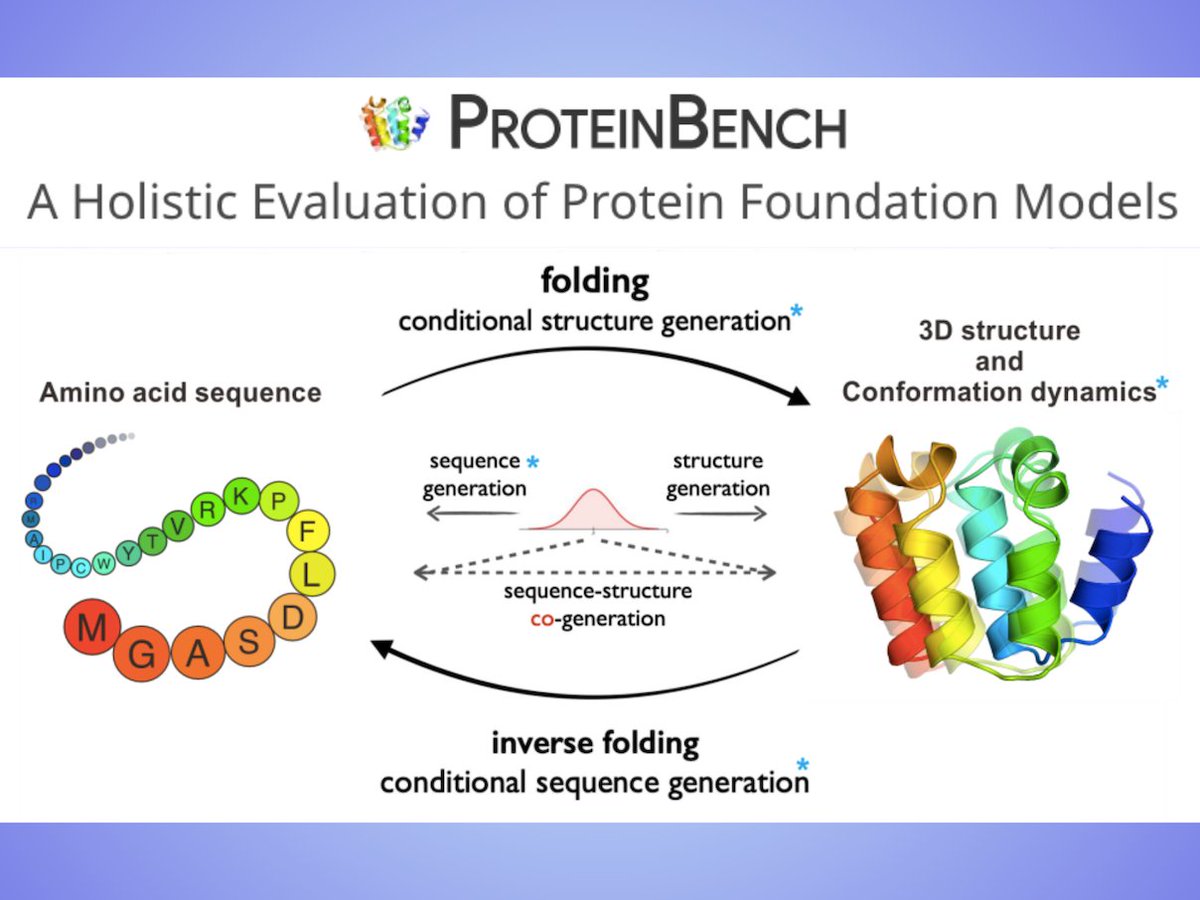
Submissions on crystal #structureprediction as well as those using lattice energy calculations as a tool to understand structure will be welcome at "Prediction of molecular crystal structures" (MS33) #ECM31Oviedo #crystallography
Learning Computational Drug Discovery and Design has not been this easy before! shoutout.wix.com/so/5aOFzm0xT?l… #drugdiscovery #moleculardocking #structureprediction #proteomics #proteins

Fresh from press #Review by Phil Bradley @UW and Brian Kuhlman @UNC on advances in protein #StructurePrediction and #ProteinDesign go.nature.com/309xj7w #ComputationalModeling #MachineLearning
Submit abtracts @ …l-biology.biochemistryconferences.com on #Molecularmodelling #Structureprediction #StructuralEnzymology #Proteomics #StructuralBiology #Geneprediction #Biochemistry #GeneSequencing #DrugDesign #HomologyModelling #Proteinengineering #NMR Explore,Learn & Share @biologyfinland

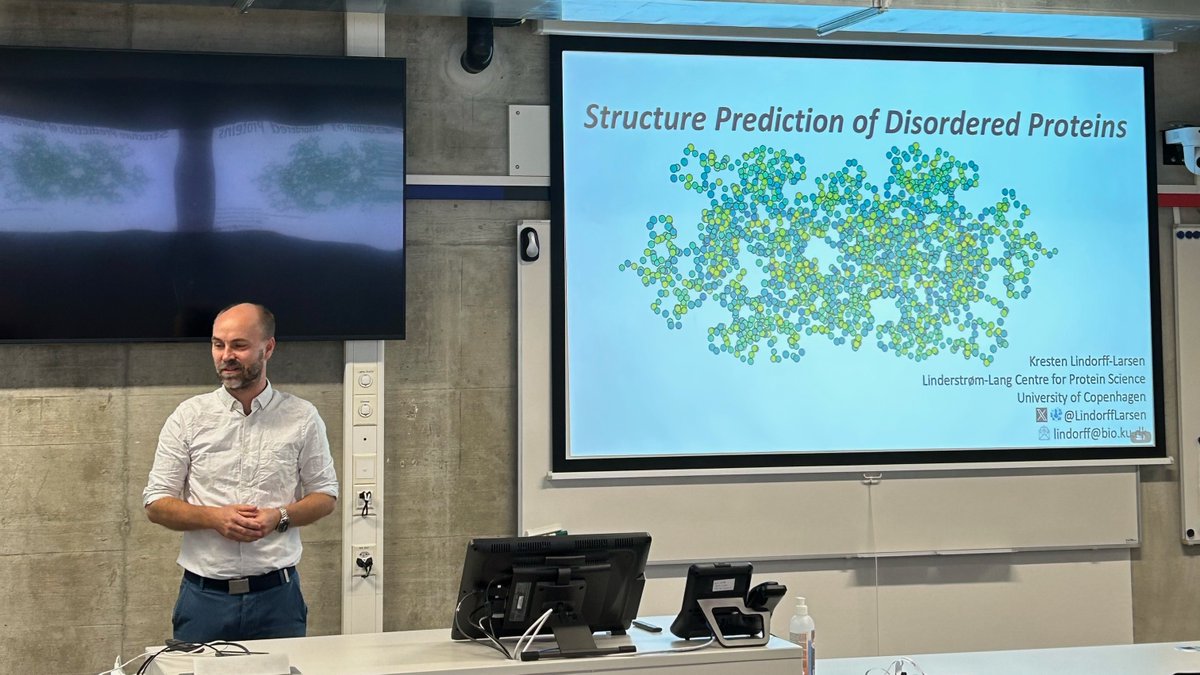
Third day of our @cecamEvents workshop "Macromolecular complexes: from ab initio and integrative modelling to functional dynamics". Today we move from #structureprediction to #proteindynamics. First speaker of the day is @LindorffLarsen from @uni_copenhagen talking about IDPs.
#MarkTuckerman @nyuniversity speaks @Cambridge_Uni bit.ly/1QM71Bn #StructurePrediction #MaterialsScience

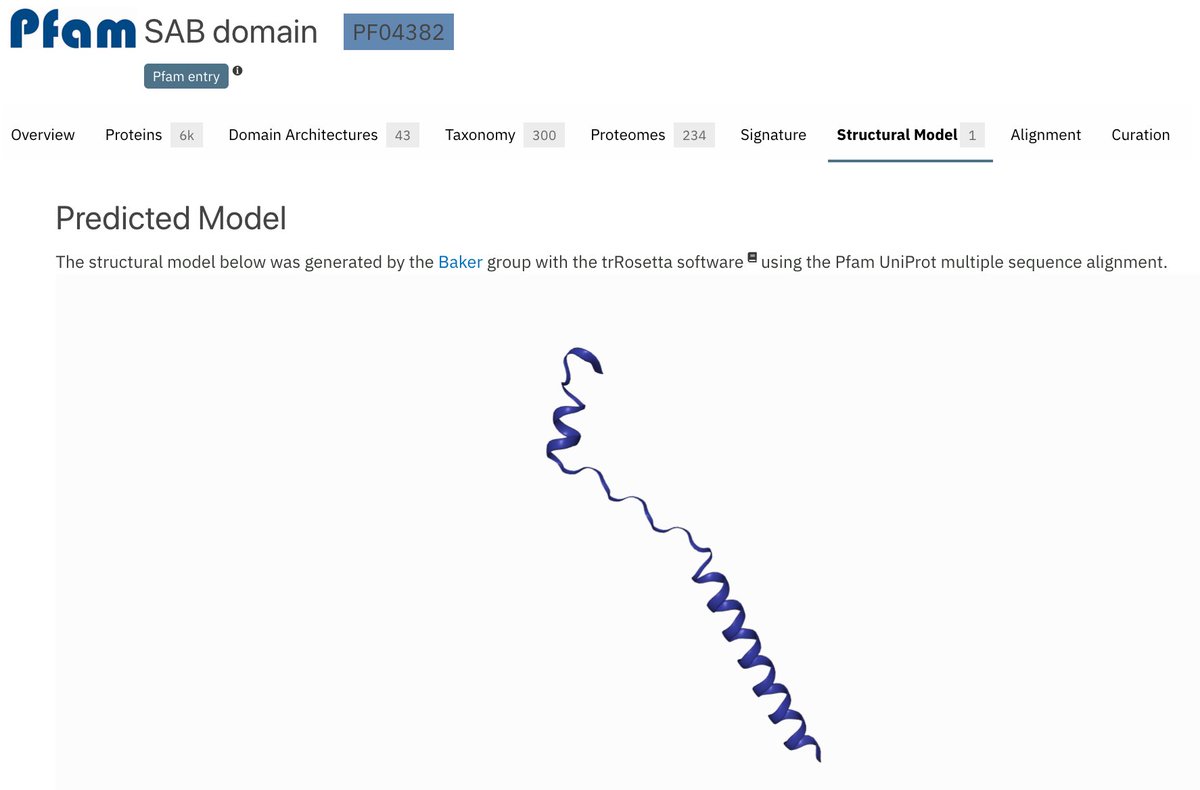
The SAB domain pfam.xfam.org/family/PF04382 doesn't really look like a domain at all. Removing its Type=Domain #structureprediction #bakermodels
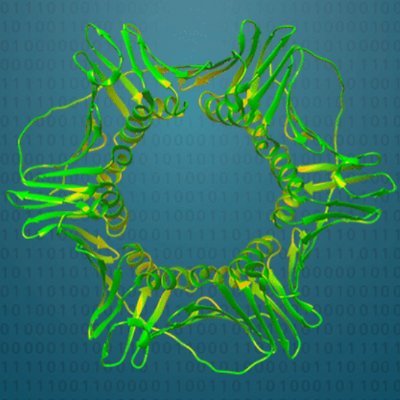
Big shout out to @interProDB for their wonderful visualization of our structure models and alignments. For example see the beautiful beta barrel of DUF1302, presumably an OMP: ebi.ac.uk/interpro/entry… #structureprediction #trRosetta #structuralbiology #deeplearning #AI

📢 New call for paper in #StructuralBioinformatics in memoriam of the work of Cyrus Chothia Edited by Alfredo Iacoangeli, Paolo Marcatili, Sarah Teichmann and Charlotte Deane Deadline: November 2020 info: fro.ntiers.in/8x9u @AlfredoIacoange @OPIGlets #StructurePrediction
🔬 Thrilled to share our group's latest preprint on implementing variable salt concentration for #RNA structure prediction in the #ViennaRNA Package! 🧬🔍 researchsquare.com/article/rs-277… #structureprediction #ViennaRNA #preprint #bioinformatics #computationalbiology
Deep learning for structure determination @IUCrJ doi.org/10.1107/S20522… @RiceUNews @RiceCompSci @RiceNatSci @RiceUIBB @ChemistryRice @WelchFoundation @NSF #BIO @AmazonScience @MSFTResearch #structureprediction #crystallography #deeplearning @IUCr #openaccess
#Protein #StructurePrediction latest
Anyone have tips for doing protein structural prediction using AlphaFold or ColabFold? I'm more interested in doing many predictions than getting super accurate ones!
Major modes of behavior within low-pLDDT regions were identified through a survey of human proteome predictions provided by the AlphaFold Protein Structure Database @ActaCrystD @IUCr #AlphaFold2 #StructurePrediction #ConditionalFolding doi.org/10.1107/S20597…

Christopher J. Williams et al.: Categorizing prediction modes within low-pLDDT regions of AlphaFold2 structures: near-predictive, pseudostructure and barbed wire #AlphaFold2 #StructurePrediction #ConditionalFolding ... #IUCr journals.iucr.org/paper?S2059798…
🧾 All structural models and evaluation data from this study are openly available via Zenodo, supporting reproducibility and further benchmarking: zenodo.org/records/148108… #OpenScience #StructurePrediction
🎁 Our February Issue is online🎁 📘“In the Limelight” issue focusing on Structural Bioinformatics. Don't miss the new Review Articles, as introduced by Guest Editors @cmslnsoares and Diana Lousa 🔓buff.ly/4aKTjeg #StructuralBioinformatics #StructurePrediction

Exploring the latest advancements in #RNA #3D #StructurePrediction, this review highlights the integration of #MachineLearning and experimental techniques in enhancing accuracy, with significant implications for drug discovery! 📄 doi.org/10.1016/j.str.… EVBC👤: @jmbujnicki
🚀 Dive into the cutting-edge world of #ProteinDesign & #StructurePrediction! We chat with David about how principles of protein engineering are reshaping the future of therapeutics. 🧬💡 Tune in to explore #DeNovoDesign and the revolutionary impact of Rosetta & RoseTTAFold!
Scientists unveil #ProteinBench—a new evaluation framework to improve transparency in protein foundation models. With its multi-metric approach, it aims to drive innovation & collaboration in 3D #structureprediction, #proteindesign, and more! Quick Read: cbirt.net/proteinbench-u…

Peptides Unfolded: Predict Structures and Discover Their Potential! Celebrate Independence Day with a 50% Discount on Our Workshop! Enroll Now: nanoschool.in/enroll-me/?pid… Know More: nanoschool.in/biotechnology/… #PeptideScience #Bioinformatics #StructurePrediction #ScienceWorkshop
Decode the Future of Peptides – Your Key to Innovation Awaits! Enroll Now: nanoschool.in/nstc/enrollmen… Know More: nanoschool.in/biotechnology/… #PeptideInnovation #BiotechWorkshop #StructurePrediction #FutureOfMedicine #DrugDiscovery #BiotechRevolution #PeptideResearch #BiotechSkills
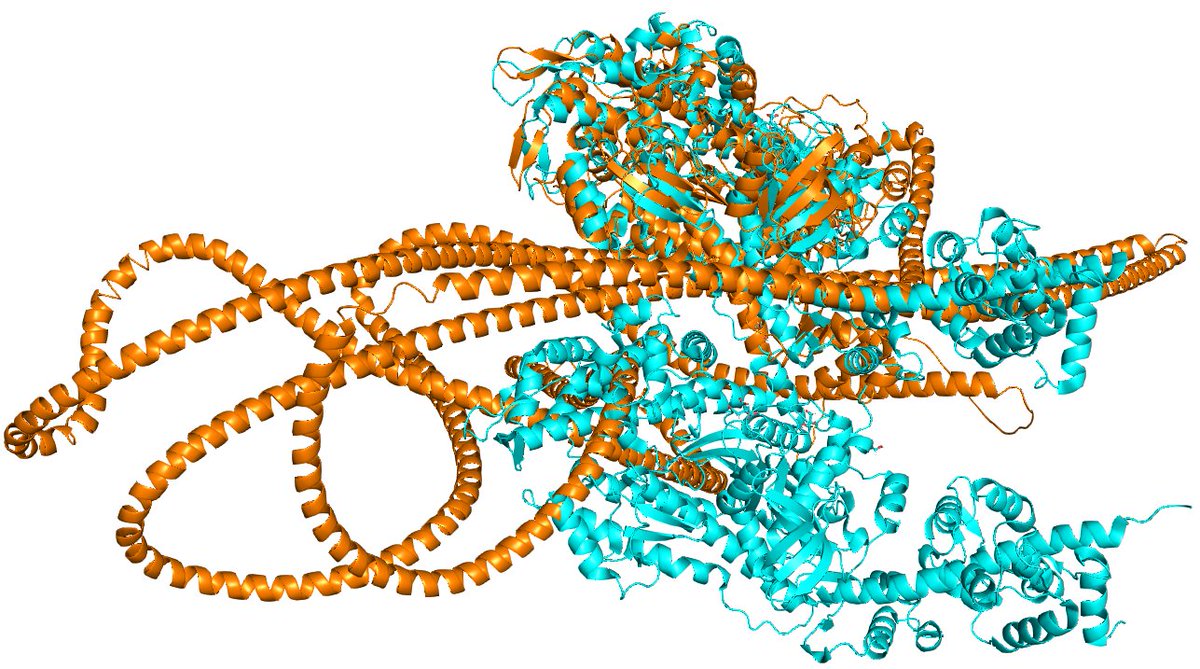
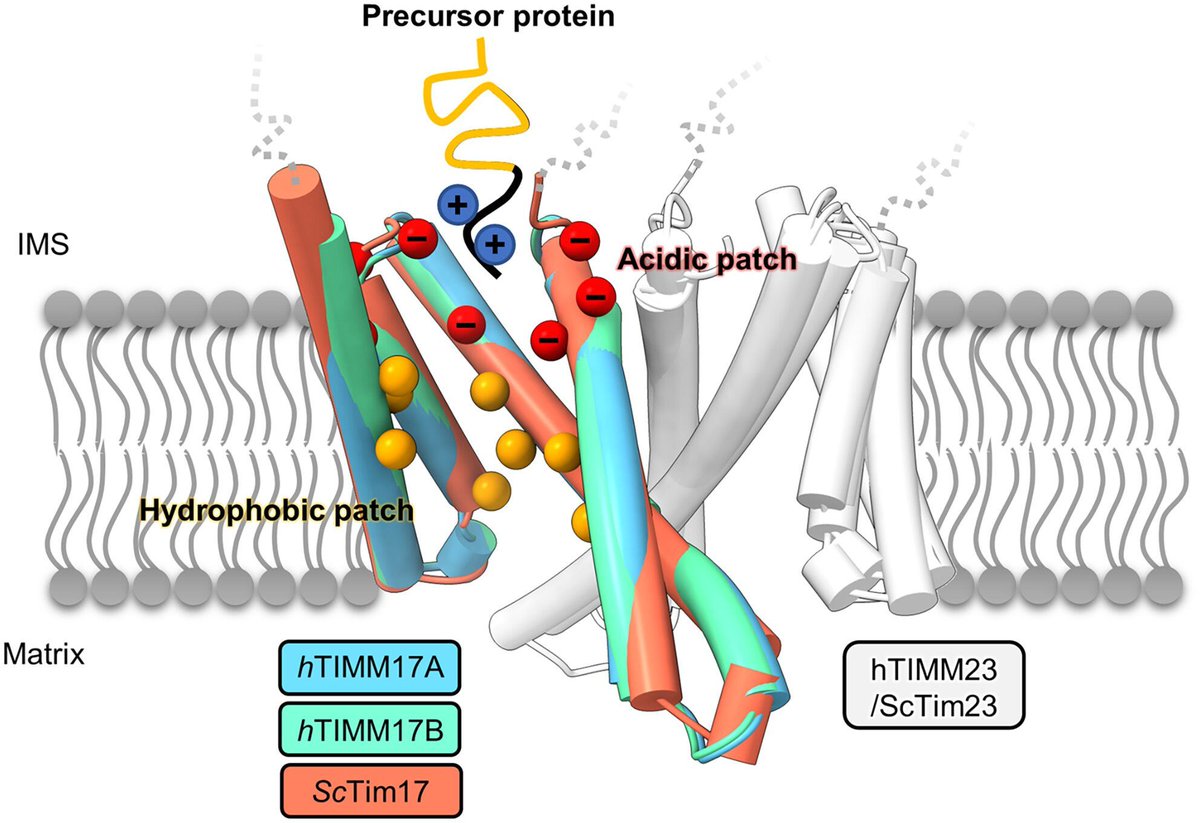
Structure prediction analysis of human core TIM23 complex reveals conservation of the protein translocation mechanism 🔗buff.ly/3VIeiaw 🥼@A_Chacinska @PDraczkowski #StructurePrediction #TIM23 #Mitochondria #StructuralBiology
Shouyang Zhang et al.: Crystallographic phase identifier of a convolutional self-attention neural network (CPICANN) on powder diffraction patterns #ComputationalModeling #StructurePrediction #XRayDiffraction ... #IUCr journals.iucr.org/paper?S2052252…
Olga Anosova et al.: The importance of definitions in crystallography #CrystalDefinition #StructurePrediction #MaterialsModeling @livuni... #IUCr journals.iucr.org/paper?S2052252…
@3dsBIOVIA Discover Studio Simulation: Harnessing AI for Predicting Structures Read More: altem.com/biovia-discove… #AI #StructurePrediction #StudioSimulation #DiscoverAI #PredictiveModeling
@3dsBIOVIA Discover Studio Simulation: Harnessing AI for Predicting Structures Read More: altem.com/biovia-discove… #AI #StructurePrediction #StudioSimulation #DiscoverAI #PredictiveModeling @Dassault3DS
#Protein #StructurePrediction latest
Anyone have tips for doing protein structural prediction using AlphaFold or ColabFold? I'm more interested in doing many predictions than getting super accurate ones!
We are delighted to announce the release of structural models for 6,370 families of unknown structure provided by Baker group @UWproteindesign: xfam.wordpress.com/2021/03/03/fol… (funded by @BBSRC) #structureprediction #trRosetta #structuralbiology #deeplearning #AI

Advances in #RNA 3D Structure Prediction #RNAstructure #StructurePrediction #compchem pubs.acs.org/doi/10.1021/ac… #vol62 #issue23 #JCIM #Reviews

Fresh from press #Review by Phil Bradley @UW and Brian Kuhlman @UNC on advances in protein #StructurePrediction and #ProteinDesign go.nature.com/309xj7w #ComputationalModeling #MachineLearning

Scientists unveil #ProteinBench—a new evaluation framework to improve transparency in protein foundation models. With its multi-metric approach, it aims to drive innovation & collaboration in 3D #structureprediction, #proteindesign, and more! Quick Read: cbirt.net/proteinbench-u…

Major modes of behavior within low-pLDDT regions were identified through a survey of human proteome predictions provided by the AlphaFold Protein Structure Database @ActaCrystD @IUCr #AlphaFold2 #StructurePrediction #ConditionalFolding doi.org/10.1107/S20597…

Learning Computational Drug Discovery and Design has not been this easy before! shoutout.wix.com/so/5aOFzm0xT?l… #drugdiscovery #moleculardocking #structureprediction #proteomics #proteins

Ok, so I decided to superimpose myosin structure to its alphafold predicted structure.. that went well.. #alphafold2 #Biochem #structureprediction

Submissions on crystal #structureprediction as well as those using lattice energy calculations as a tool to understand structure will be welcome at "Prediction of molecular crystal structures" (MS33) #ECM31Oviedo #crystallography

📢 New call for paper in #StructuralBioinformatics in memoriam of the work of Cyrus Chothia Edited by Alfredo Iacoangeli, Paolo Marcatili, Sarah Teichmann and Charlotte Deane Deadline: November 2020 info: fro.ntiers.in/8x9u @AlfredoIacoange @OPIGlets #StructurePrediction

Structure prediction analysis of human core TIM23 complex reveals conservation of the protein translocation mechanism 🔗buff.ly/3VIeiaw 🥼@A_Chacinska @PDraczkowski #StructurePrediction #TIM23 #Mitochondria #StructuralBiology

The SAB domain pfam.xfam.org/family/PF04382 doesn't really look like a domain at all. Removing its Type=Domain #structureprediction #bakermodels

Submit abtracts @ structuralbiology.conferenceseries.com on #Molecularmodelling #Structureprediction #StructuralEnzymology #Proteomics #StructuralBiology #Geneprediction #Biochemistry #GeneSequencing #DrugDesign #HomologyModelling #Proteinengineering #NMR Explore, Learn & Share @structuralbiol1

#MarkTuckerman @nyuniversity speaks @Cambridge_Uni bit.ly/1QM71Bn #StructurePrediction #MaterialsScience

Submit abtracts @ …l-biology.biochemistryconferences.com on #Molecularmodelling #Structureprediction #StructuralEnzymology #Proteomics #StructuralBiology #Geneprediction #Biochemistry #GeneSequencing #DrugDesign #HomologyModelling #Proteinengineering #NMR Explore,Learn & Share @biologyfinland

Big shout out to @interProDB for their wonderful visualization of our structure models and alignments. For example see the beautiful beta barrel of DUF1302, presumably an OMP: ebi.ac.uk/interpro/entry… #structureprediction #trRosetta #structuralbiology #deeplearning #AI

Deep learning for structure determination @IUCrJ doi.org/10.1107/S20522… @RiceUNews @RiceCompSci @RiceNatSci @RiceUIBB @ChemistryRice @WelchFoundation @NSF #BIO @AmazonScience @MSFTResearch #structureprediction #crystallography #deeplearning @IUCr #openaccess

Third day of our @cecamEvents workshop "Macromolecular complexes: from ab initio and integrative modelling to functional dynamics". Today we move from #structureprediction to #proteindynamics. First speaker of the day is @LindorffLarsen from @uni_copenhagen talking about IDPs.

🎁 Our February Issue is online🎁 📘“In the Limelight” issue focusing on Structural Bioinformatics. Don't miss the new Review Articles, as introduced by Guest Editors @cmslnsoares and Diana Lousa 🔓buff.ly/4aKTjeg #StructuralBioinformatics #StructurePrediction

Something went wrong.
Something went wrong.
United States Trends
- 1. Aaron Gordon 20.2K posts
- 2. Steph 46.1K posts
- 3. Jokic 21.1K posts
- 4. Wentz 24.6K posts
- 5. Vikings 51.5K posts
- 6. Warriors 72.1K posts
- 7. #criticalrolespoilers 9,497 posts
- 8. #DubNation 3,962 posts
- 9. Halle 16.9K posts
- 10. #EAT_IT_UP_SPAGHETTI 191K posts
- 11. Chargers 56.4K posts
- 12. #LOVERGIRL 15.1K posts
- 13. Pacers 22.2K posts
- 14. Cam Johnson 1,540 posts
- 15. Nuggets 23.2K posts
- 16. Hobi 34.9K posts
- 17. Shai 23.5K posts
- 18. Brosmer 3,827 posts
- 19. SPAGHETTI FT J-HOPE OUT NOW 49.5K posts
- 20. Herbert 16.6K posts