Alan Boyle
@AP_Boyle
Genomics/Bioinformatics Assoc. Prof., University of Michigan Msstate, Duke, Stanford, Michigan Genetic Variation, Silencers, RegulomeDB, Nanopore Tech
내가 좋아할 만한 콘텐츠
Check out our study on using donor-specific assembly to facilitate the discovery of somatic genetic variants in bulk and single neurons of the postmortem brain. Colab with @ryan_e_mills @AP_Boyle @mikemc43 @CamilleMumm @YanmingGan
Critical clues are being revealed about the impact of somatic mutations on human health and disease. A SMaHT study demonstrates how personalized assemblies and single-cell sequencing can sharpen the detection of somatic mutations in the human brain 🧠. biorxiv.org/content/10.110…
1/ I am beyond thrilled to share our new paper in @CellCellPress! We explored the regulatory principles underlying pleiotropy in psychiatric disorders. cell.com/cell/abstract/…
Our ChromBPNet preprint out! Huge congrats to @panushri25! This was quite a slog for both of us but we r very proud of this one! It is a long read but worth it IMHO. Methods r in the supp. materials. Link in the next tweet. Bluetorial coming soon .. 1/
MNI is actively recruiting for 𝙨𝙚𝙫𝙚𝙧𝙖𝙡 𝗳𝗮𝗰𝘂𝗹𝘁𝘆 𝗽𝗼𝘀𝗶𝘁𝗶𝗼𝗻𝘀, seeking the best and brightest in #neuroscience to fill these roles. Please 💛 and r/t this to help us reach some great candidates! 〽️🧠 More info: michmed.org/GqG8z

Excited to share the white paper for the IGVF Consortium, supported by @genome_gov Deciphering the impact of genomic variation on function rdcu.be/dSZdm
Understanding the function of an entire genome is no easy task! Most genes likely have more than one function. To understand the functional effects of genomic variation, NHGRI launched a consortium called Impact of Genomic Variation on Function (IGVF). genome.gov/news/IGVF

New paper from my postdoc work at @UMichResearch is now out in @ScienceAdvances 🎉 very proud to share the culmination of nearly 4 years of hard work with a fantastic group of colleagues and collaborators. A 🧵 of what we found out about RFC1/CANVAS 👉 science.org/doi/10.1126/sc…
Our first study in a new repeat expansion disorder: CANVAS. @connor_maltby used patient iPSNs to define how this non-reference AAGGG repeat impacts neuronal function. He finds a novel role for the repeat in disease that is independent of RFC1 function science.org/doi/10.1126/sc…
Systematic investigation of allelic regulatory activity of schizophrenia-associated common variants hubs.li/Q02v7f3S0 @hyejung_won @jsteinlab @srikosuri @mikelove @_josedavila @brad_ruzicka @AP_Boyle Up-and-coming genomics research from @CellGenomics #neurogenomics
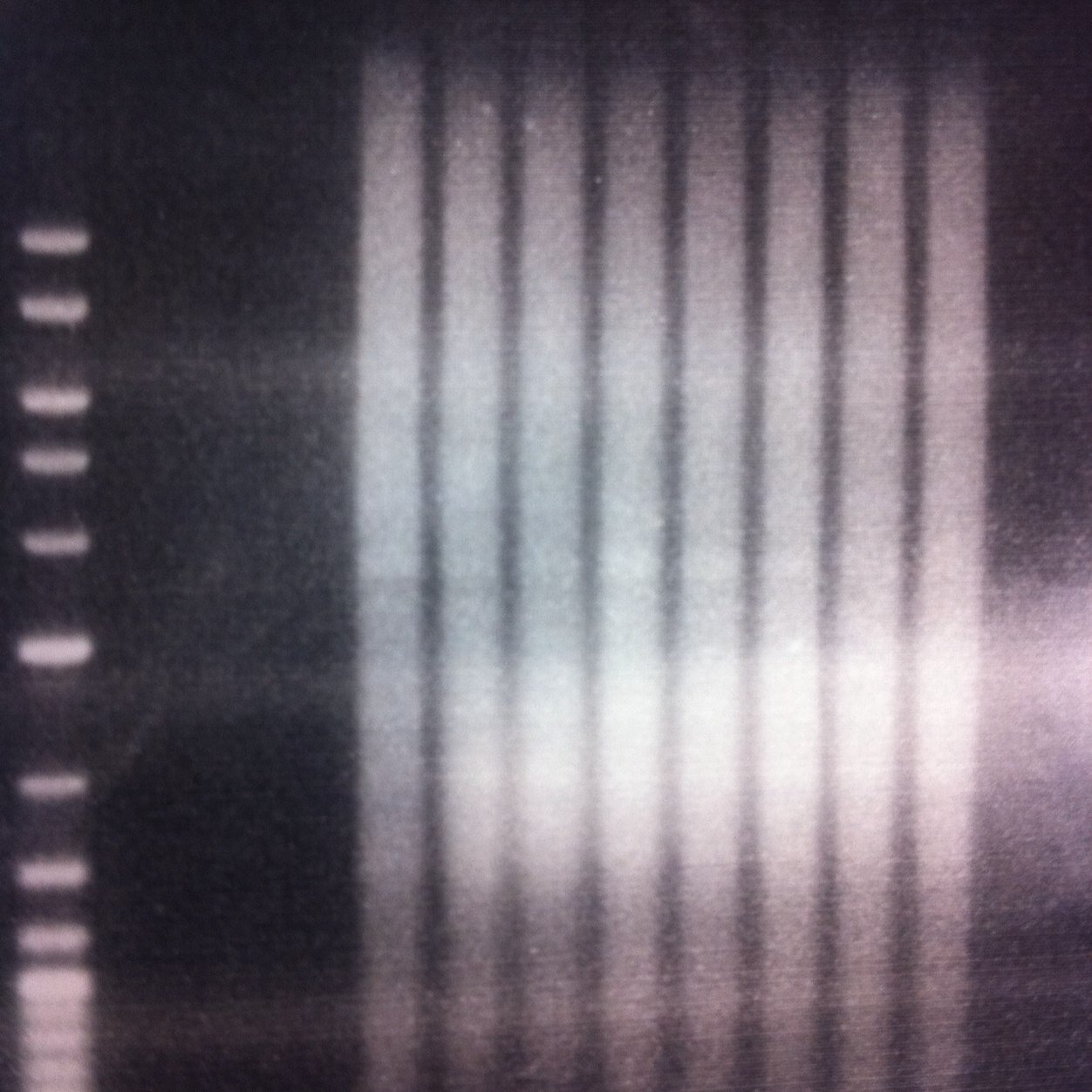
New @AP_Boyle and @realToddLab collaboration is now up on @medrxivpreprint 🥳 we used targeted long read sequencing and HMMSTR to successfully identify and genotype known or suspected large tandem repeat expansions at many loci in patient derived samples. medrxiv.org/content/10.110…
Our preprint is out! We hacked the @nanopore sequencer to read amino acids and PTMs along protein strands. This opens up the possibility for barcode sequencing at the protein level for highly multiplexed assays, PTM monitoring, and protein identification! biorxiv.org/content/10.110…
I am beyond excited to share our new paper on massively parallel reporter assays on schizophrenia-associated variants. This is in collaboration with @jsteinlab, @srikosuri, @mikelove, @_josedavila, @brad_ruzicka, @AP_Boyle. 🧵 follows.
Systematic investigation of allelic regulatory activity of schizophrenia-associated common variants dlvr.it/SwBgpY
Current tools can only partially validate plasmid sequences due to their inability to resolve complex regions. In this interview, Mel Englund discussed how nanopore sequencing can span entire plasmids allowing for accurate validation of complete plasmids: bit.ly/3KSYj4V
Current methods lack the scalability to validate full-length plasmid sequences. In this interview learn how nanopore technology provides a fully scalable solution for full-length plasmid validation, from the portable MinION to the high-throughput GridION. bit.ly/3QOiMeM
Want to join a fun, supportive, and intellectually curious lab that explores the interface between fundamental mRNA biology and human neurological disease? If so, the Todd lab might be a good fit. Check out our posting below or visit our website!

US citizens or permanent residents from an under-represented group! High school thru postdoc/faculty. You can contact any NIH-funded researcher you admire & suggest applying for this supplement to join their lab! Many, like me, are happy to host! grants.nih.gov/grants/guide/p…
📢New in @HGGAdvances! @AP_Boyle & colleagues explore the challenges posed when screening for de novo variants in complex phenotypes bit.ly/43f86ZE

Really excited to be part of this with @AP_Boyle and @mikemc43! Our part of the project will be focused on exploring somatic mosaicism in single cells as well as targeted capture of specific types of variation in bulk tissue using long read nanopore sequencing.
News: NIH launches $140 million effort to investigate genetic variation in normal human cells and tissues dlvr.it/SntWlH
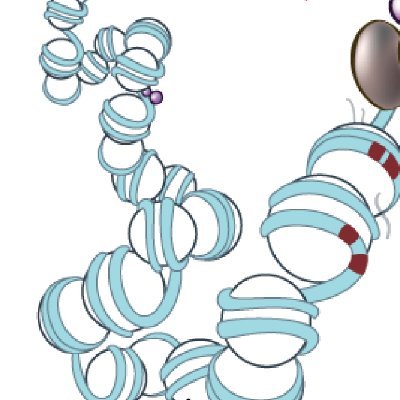
Happy to share that our Michigan #CASP15 team ended up placing second in Regular Targets, first in Multimeric Targets, and first in Inter-domain prediction (1/3)

United States 트렌드
- 1. Prince Andrew N/A
- 2. Andrew Mountbatten N/A
- 3. Good Thursday N/A
- 4. Happy Friday Eve N/A
- 5. Sandringham N/A
- 6. $TOTO N/A
- 7. Cigar N/A
- 8. Perú N/A
- 9. Board of Peace N/A
- 10. Strickland N/A
- 11. Reese N/A
- 12. Acuff N/A
- 13. Josh Hubbard N/A
- 14. Swerve N/A
- 15. $WAR N/A
- 16. #PoppyPlaytimeChapter5 N/A
- 17. Glenn Beck N/A
- 18. Darryn Peterson N/A
- 19. Prototype N/A
- 20. Hangman N/A
Something went wrong.
Something went wrong.