BioDataScience Lab
@BioDataSc
We apply our expertise in #Statistics and #ML to help answer principal questions in #microbiology and #microbiome. Part of @LMU_Muenchen and @HelmholtzMunich.
You might like
🎉 Had a blast at #ISMBECCB2023 last week! So many inspiring talks, awesome posters and... stairs in Lyon 🇫🇷😃 Excited to put all the amazing ideas we came up with into action!💡🚀 #microbiome #datascience @HelmholtzMunich @LMU_Muenchen

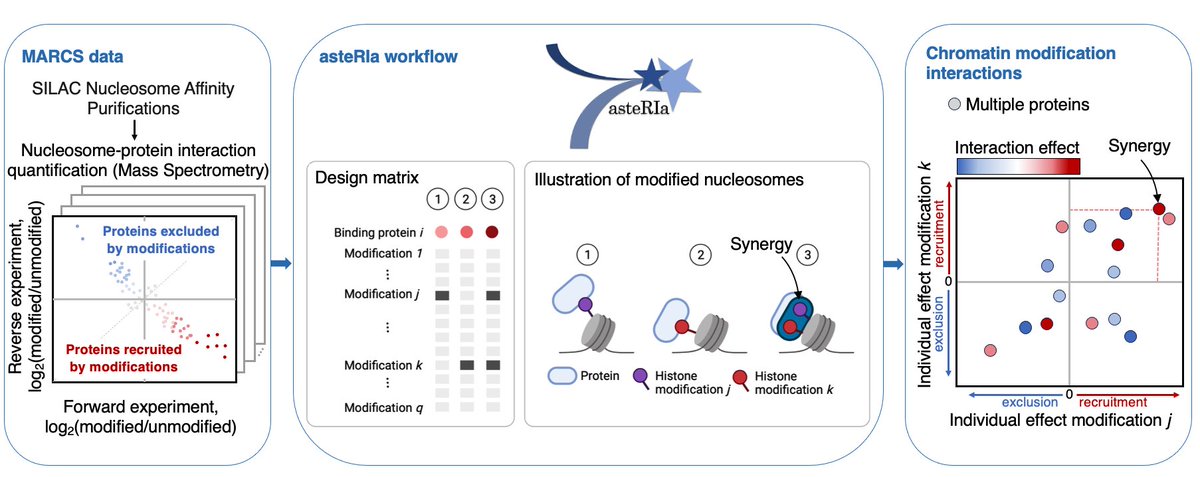
I'm excited to share that our paper, 'asteRIa enables robust interaction modeling between chromatin modifications and epigenetic readers,' is now out in NAR! academic.oup.com/nar/article/do… Through statistics and large-scale proteomics data, we disentangle combinatorial epigenetics 👇
Considering microbial interactions in prediction tasks is crucial! 🦠📊 Excited about our collaboration with @microbionaut and Jacob Bien. Check out our pre-print: doi.org/10.1101/2024.0…
Had an awesome time presenting our work at #EESAIBio! Feel free to come up and chat and check out our pre-print! biorxiv.org/content/10.110…

Excited to share our work "asteRIa enables robust interaction modeling between chromatin modifications and epigenetic readers", where we utilize the MARCS data for deeper insights into combinatorial epigenetics. Great collab between Bartke lab and @BioDataSc at @HelmholtzMunich
Chromatin modifications wield immense power in gene regulation! 🧬 Check out this amazing work just published in @Nature. 🔍 #ChromatinModifications #GeneRegulation
Check out this amazing data resource to study how chromatin modifications regulate protein binding behavior by Bartke lab and @robert_ife 🧬 I'm very proud to be part of this project 👇
📊 Unveiling Stable Interactions in High-Throughput data! 💡@StadlerMara introduce #robust #statistical workflow: 🔗 Application to interaction effects between chromatin modifications 🧬 Lasso model for hierarchical interactions 🔎 Stability-based model selection #ISMBECCB2023

🔬✨ Introducing MolE! Join @Scietwas to witness MolE's empowering chemical compound #analysis 🌐🌟 ⚙️ Molecular representation learned through embedding decorrelation ✨ Predicted growth-inhibitory effects against human gut pathogens #ML #antimicrobial #resistance #ISMBECCB2023

🔍Discover the power of PIMs (compositional power interaction models) in handling @HiTSeq data! 🧬 @JohannesOstner showcases PIMs' capabilities in DA testing with correlated features while respecting zero inflation and compositional constraints 📈🎯 #ISMBECCB2023

🌐Join Viet Tran in Overcoming Statistical Challenges in Microbiome and Single Cell Research! 🏆🧪 📈Innovative Two-Stage Workflow: 1️⃣Hierarchical grouping of microbial or cell-type features 2️⃣Explicit handling of non-compositional covariates #microbiome #singlecell #ISMBECCB2023

🔬🌿 Reproducible #microbiome #dataanalysis with #QIIME2 🐍 #python! 🔍 q2-gglasso: Sparse microbial network estimation 📈 q2-classo: Sparse log-contrast regression and classification Join @ovlasovets @microbiome_cosi to hear more about #openscience and #softwaredevelopment🚀💻

🦠Unlocking Hierarchical Relationships in Viral #Metagenomic Data with #VirNest! 🧬 1️⃣ Virus gene-sharing network estimation by leveraging hierarchical protein association 2️⃣ Hierarchical partitions through nested stochastic block model Join @DanielePugno in unveiling #microbiome

Discover the power of #NetCoMi, the #R package disentangling microbial association #networks! 📊🔬 🦠 Understand complex #microbial interactions 🌱 Statistical network estimation made easy 🔍 Unveil the differences between groups Don't miss a showcase made by @stefpeschel ⭐

🔬 Today was an incredible day at @deng_lab! Huge thanks to @JinlongRu, @KhanMirzaei, @Scietwas, @soph_eliza94, and @DanielePugno for sharing their expertise. Looking forward to new collaborations and more exciting discussions in the future🙌 #virome #microbiome @HelmholtzMunich

That went by quickly! Already starting the last week of my three-month research stay at @USCMarshall. Together with Jacob Bien and @microbionaut, we explored how to model compositional high-throughput sequencing data in a meaningful way. Stay tuned!


Our lab @IEM_Augsburg in Universität Klinikum #Augsburg. We SAW where skin #microbiome data comes from, and now will think twice before "df.dropna()" any samples🙀. Big thanks to @amedeodetomassi for an amazing lab tour, Luise Rauer and their colleagues for making it possible😎👍

We @BioDataSc have open positions for PhD/Postdocs at the interface of statistics, optimization, and microbiome data analysis #stats, #optimization, #microbiome, #datascience, #oceanmicrobiome #gutmicrobiome @CompHealthMuc @LMU_Muenchen; PM me or send an email for more info!!
Please, meet @Scietwas (Roberto), @SharmaLab1, @AnaRitaBrochad1 and Victoria - a data generation 🤖 from the clip. They expect to have data for over 200k experiments for #StressRegNet project which is a part of @bayresq ’s effort to tackle #antibiotic resistance💊💪.
United States Trends
- 1. Nancy Pelosi 74.4K posts
- 2. Marshawn Kneeland 47.2K posts
- 3. Ozempic 7,242 posts
- 4. Craig Stammen 1,949 posts
- 5. Michael Jackson 73.1K posts
- 6. Gordon Findlay 3,193 posts
- 7. Oval Office 26.2K posts
- 8. Pujols N/A
- 9. #MichaelMovie 69.1K posts
- 10. GLP-1 5,299 posts
- 11. Jaidyn 1,825 posts
- 12. Kyrou N/A
- 13. Abraham Accords 5,458 posts
- 14. Novo Nordisk 7,599 posts
- 15. Kazakhstan 7,238 posts
- 16. #NO1ShinesLikeHongjoong 38.5K posts
- 17. Unplanned 9,389 posts
- 18. #영원한_넘버원캡틴쭝_생일 37.7K posts
- 19. #ChelleyxEOS 1,718 posts
- 20. Baxcalibur 6,535 posts
Something went wrong.
Something went wrong.