ForliLab
@ForliLab
Molecular Modeling & Drug Design. The lab of Stefano Forli at Dept. of Integrative Structural & Computational Biology @scrippsresearch Home of the AutoDock
Вам может понравиться
Important Update: we've decided to close our Twitter account because the increasing censorship and political controversies do not align with our values. You can continue following us on @forlilab.bsky.social. We will not post anymore here.
Transthyretin’s structural asymmetry & “molecular breathing” drive its misfolding in ATTR—a discovery by Prof. Gabriel Lander (@LanderLab) & Prof. Jeff Kelly, published in @NatureSMB, that could inform new therapies for amyloid diseases. More: ow.ly/xCx850UVbbl

Kudos @ForliLab!!!
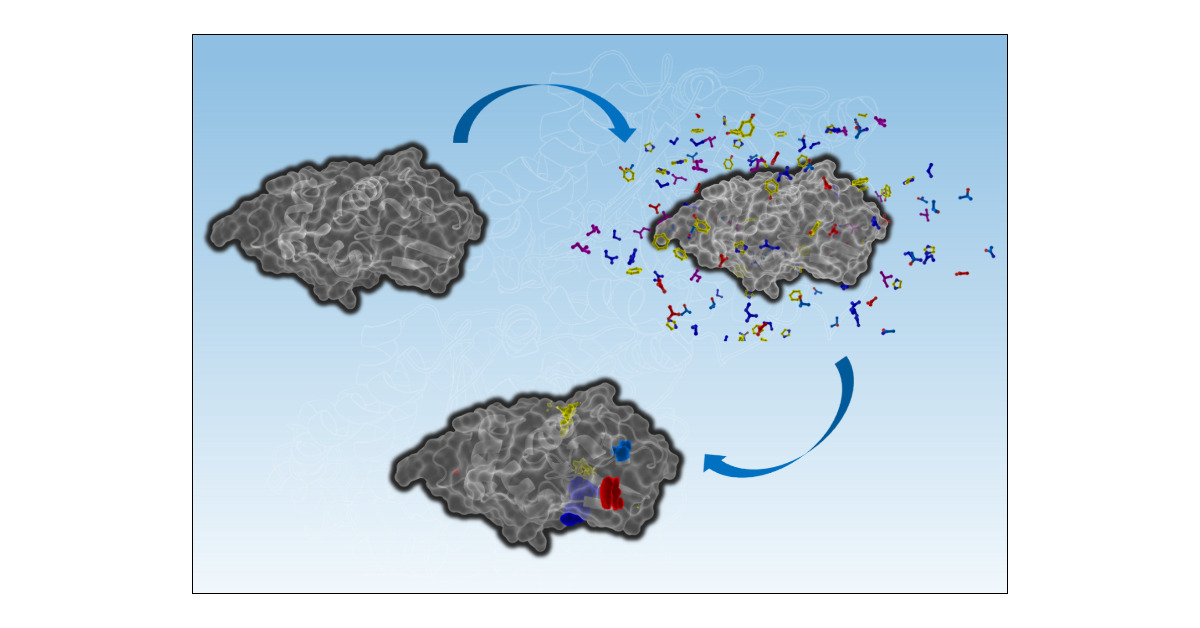
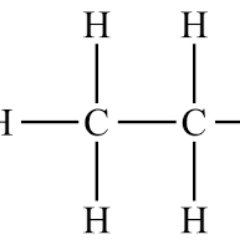
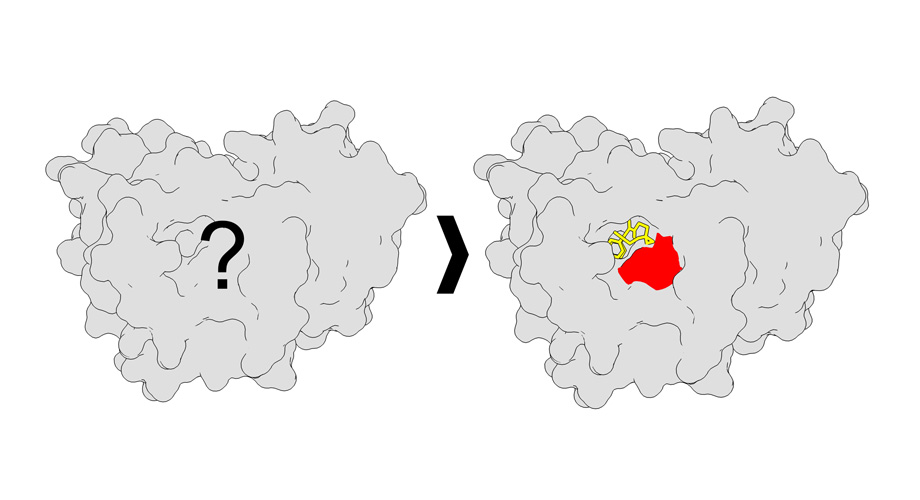
#HighlightOfTheWeek Cosolvent molecular dynamics are an increasingly popular form of MD simulations where small molecule cosolvents are added to the system, helping to identify cryptic and allosteric pockets in proteins. #compchem pubs.acs.org/doi/10.1021/ac…
Congratulations to Dr. @Batuujin for successfully defending his thesis yesterday! We are so proud of you and your work in our lab and in @WardLab1 . We can only look forward to the amazing science you will continue to do! #otop #sour_zebrafish

If you’re interested in fast sampling and flexible and automated creation of cosolvent systems with @openmm_toolkit check this out! Huge thanks to the team @je_eberhardt @diogo_stmart @manuco17 @moonii1020 @Joe_r_loeffler @ForliLab pubs.acs.org/doi/10.1021/ac… ⚛️🧬
AlphaFold2+MD simulations = powerful insights into protein conformational ensembles, but can it capture all experimental structural variations? Great collaborative effort!! @Structure_CP @RRiccabona @OPIGlets @scrippsresearch @DTUtweet #compchem sciencedirect.com/science/articl…
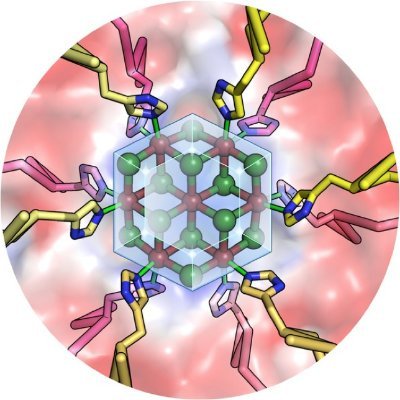
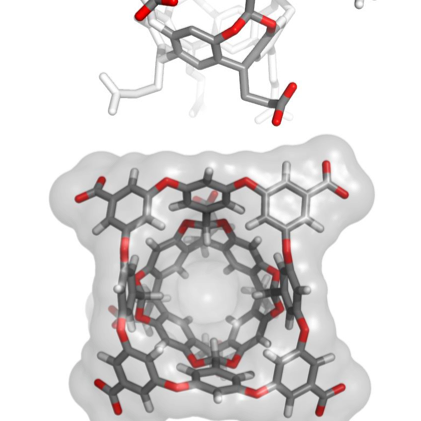
Mesoscale Explorer molstar.org/me/ Paper out !! Excellent work with @asrmoin @dsgoodsell and @sehnaldavid doi.org/10.1002/pro.51…
Our second years @AllisonBarkdull, @YutingZhan8328, and @Renhao4 received lab coats this weekend! #computationalscientists #whataretheygonnadowithlabcoats

congratulations to Ishan for winning second place for his talk at Scripps' Graduate Research Symposium #IshanFold

Do you know Python and have experience with RDKit? Do you want to work in an exciting and multidisciplinary environment?The AutoDock project is hiring a programmer, apply here! recruiting2.ultipro.com/SCR1003TSRI/Jo…
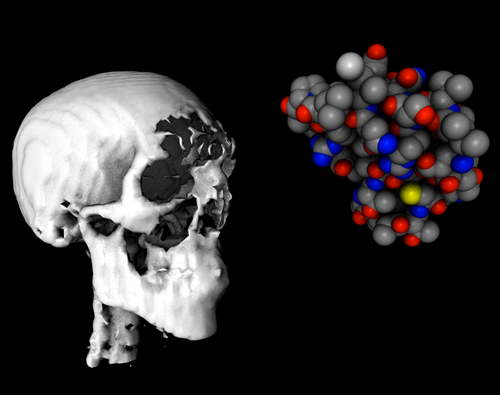
A new @nchembio study, led by @_chrisgparker_ alongside Jacob M. Wozniak and Weichao Li, created a high-resolution technique for finding new druggable sites for any human disease, from cancer to Alzheimer’s disease. More at: ow.ly/9NLN50Qnb7x
United States Тренды
- 1. Good Monday 31.9K posts
- 2. #MondayMotivation 26.8K posts
- 3. Victory Monday N/A
- 4. Jamaica 63.9K posts
- 5. Category 5 16.5K posts
- 6. #MondayMorning 1,030 posts
- 7. Hurricane Melissa 39.7K posts
- 8. Tomlin 13.4K posts
- 9. Austin Reaves 51.1K posts
- 10. #BreachLAN2 3,700 posts
- 11. GameStop 55.2K posts
- 12. South China Sea 18.1K posts
- 13. Zayne 26.9K posts
- 14. Milei 578K posts
- 15. Talus Labs 18.2K posts
- 16. Hochul 21.5K posts
- 17. Dolly 13.2K posts
- 18. Tanzania 59.4K posts
- 19. Derry 22.6K posts
- 20. Ron Paul N/A
Вам может понравиться
-
 ACS COMP
ACS COMP
@ACSCOMP -
 William L. Jorgensen
William L. Jorgensen
@JorgensenWL -
 Gianni De Fabritiis
Gianni De Fabritiis
@gdefabritiis -
 Sofia Kiriakidi
Sofia Kiriakidi
@sofiak1r -
 Giulia Palermo
Giulia Palermo
@palermo_lab -
 David Mobley
David Mobley
@davidlmobley -
 Ariane Nunes Alves
Ariane Nunes Alves
@anunesalves -
 Zoe Cournia
Zoe Cournia
@zoecournia -
 Cole Group
Cole Group
@ColeGroupNCL -
 BindingDB
BindingDB
@BindingDb -
 pen(Taka)
pen(Taka)
@iwatobipen -
 Dr. Javier GM.
Dr. Javier GM.
@JaviChem_
Something went wrong.
Something went wrong.