The Gilbert Lab
@GilbertLabRNA
We love RNA
You might like
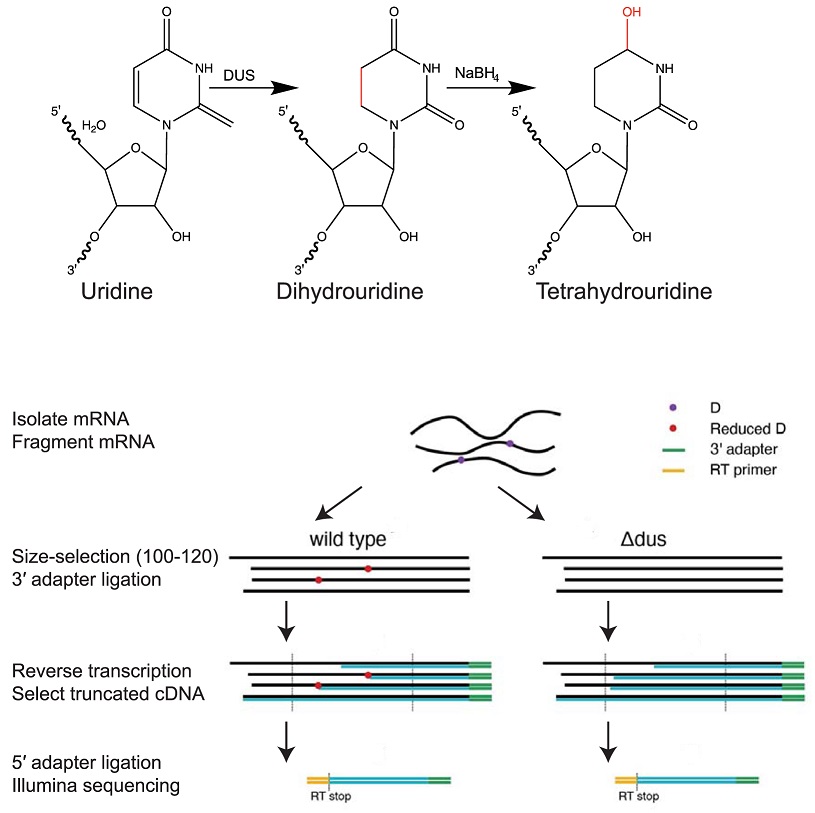
We are so excited to share our latest work profiling dihydrouridines across the yeast transcriptome! 🎉🎉 Led by grad student extraordinaire @a_draycott, this was a wonderful collaboration with fellow C-wing RNA enthusiasts @LeoSchaerf and @NeugebauerKarla journals.plos.org/plosbiology/ar…
Congratulations to our rock star grad students Cassandra and @Cole_JT_Lewis who both won poster awards at #RNA22! 🎉🎉🎉
Gilbert labbies past and present! So wonderful to see everyone again. Now we’re just waiting on the dancing 💃🏻 🕺🏻 #RNA2022 #RNA22

Novel method (D-seq) for transcriptome-wide high-res mapping of dihydrouridine reveals this #RNAmodification at new locations across the yeast transcriptome, suggesting broad role in folding functional RNA structures @a_draycott @GilbertLabRNA #PLOSBiology plos.io/3wR5Mta
Congratulations @roniederer!! We can’t wait to see what cool new discoveries you make about translational control! 🎉
In a span of < 1 wk, @GilbertLabRNA publishes 2 incredible @ACS_Research stories-1st w insights into how mRNAs are born, tumors & mets; & then @AmericanCancer PF @roniederer publishes cell.com/cell-systems/f… showing how these mRNA instructions are used, & how to manipulate them🤯
More good news! 🎉 Here we uncover roles for pseudouridylation and pseudouridine syntheses in alternative mRNA processing. Spearheaded by the fantastic @nicolemmart, this was a collaborative effort with @yeo_lab Congratulations to all authors! sciencedirect.com/science/articl…
More good news! 🎉 Here we uncover roles for pseudouridylation and pseudouridine syntheses in alternative mRNA processing. Spearheaded by the fantastic @nicolemmart, this was a collaborative effort with @yeo_lab Congratulations to all authors! sciencedirect.com/science/articl…
Check out the latest work from our lab! Congratulations to @roniederer @bzinsh and @mfrd18! 🎉🎉🎉 authors.elsevier.com/c/1eR4-8YyDffJ…
Excited that the Martinez Lab @StanfordMed @ChemSysBio @Stanford_ChEMH @DevBioStanford is hiring! We are looking for a Research Assistant to work on projects related to how RNA modifications and mRNA processing control gene expression. Apply - job ad here: careersearch.stanford.edu/jobs/life-scie…
Huge congratulations to @Cole_JT_Lewis for getting the NOA on his F31 this week!!! #sweet16 🎊🍾🎉🎊🍾🎉

Oh hey we know that paper 😄
bioRxiv: Cellular translational enhancer elements that recruit eukaryotic initiation factor 3 dlvr.it/S3x43V
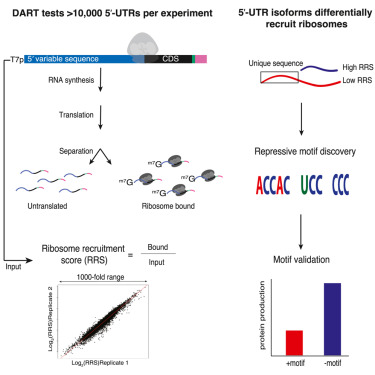
We are delighted to share our latest work on translational control elements that bind eIF3! biorxiv.org/content/10.110…
Huge congratulations to @nicolemmart who is starting her own lab @Stanford! Keep an eye out for exciting work on #RNA processing and modifications!
I am delighted to announce that the Martinez Lab is starting in January 2022 @ChemSysBio @DevBioStanford @Stanford_ChEMH! I am so grateful to my amazing mentors @WendyGScientist and Kristen W Lynch and to @GilbertLabRNA for their unconditional support.

Starting very soon!
I am delighted to announce that the Martinez Lab is starting in January 2022 @ChemSysBio @DevBioStanford @Stanford_ChEMH! I am so grateful to my amazing mentors @WendyGScientist and Kristen W Lynch and to @GilbertLabRNA for their unconditional support.

It's out! Dr. Yu Sun in the lab identified merafloxacin as a pan-betacoronavirus programmed -1 ribosomal frameshift inhibitor. Great collaboration with @BrettLindenbach @WendyGScientist @roniederer @WilenLab @HoLabHIV and YCMD colleagues @YaleWestCampus pnas.org/content/118/26…
Great science with great people!
I am delighted to announce that the Martinez Lab is starting in January 2022 @ChemSysBio @DevBioStanford @Stanford_ChEMH! I am so grateful to my amazing mentors @WendyGScientist and Kristen W Lynch and to @GilbertLabRNA for their unconditional support.

Don’t miss @roniederer sharing her work identifying novel translational control elements during the mechanisms and regulation of translation session this morning at #RNA2021!
United States Trends
- 1. #GMMTV2026 1.78M posts
- 2. MILKLOVE BORN TO SHINE 296K posts
- 3. Good Tuesday 24.9K posts
- 4. WILLIAMEST MAGIC VIBES 46K posts
- 5. #tuesdayvibe 1,822 posts
- 6. Barcelona 153K posts
- 7. TOP CALL 9,400 posts
- 8. AI Alert 8,184 posts
- 9. Alan Dershowitz 2,971 posts
- 10. Barca 80.2K posts
- 11. Moe Odum N/A
- 12. Check Analyze 2,435 posts
- 13. Token Signal 8,604 posts
- 14. Brock 42.8K posts
- 15. Purdy 28.7K posts
- 16. Unforgiven 1,145 posts
- 17. Market Focus 4,653 posts
- 18. Enemy of the State 2,531 posts
- 19. Bryce 21.5K posts
- 20. Dialyn 8,262 posts
You might like
-
 The RNA Society
The RNA Society
@RNASociety -
 UMich RNA Center 〽️🧬
UMich RNA Center 〽️🧬
@umichrna -
 Jr RNA Scientists
Jr RNA Scientists
@jrRNAscientists -
 RNA
RNA
@RNAJournal -
 Barna Lab
Barna Lab
@mbarnalab -
 RNA Therapeutics Institute
RNA Therapeutics Institute
@RTI_UMassChan -
 Nils Walter Lab
Nils Walter Lab
@NilsWalterLab -
 Cambridge RNA Club
Cambridge RNA Club
@CambridgeRNA -
 Woodson Lab
Woodson Lab
@woodson_lab -
 Schraga Schwartz
Schraga Schwartz
@SchragaSchwartz -
 Jinwei Zhang Lab
Jinwei Zhang Lab
@jinweizhang -
 The RNA Institute
The RNA Institute
@TheRNAInstitute -
 Christine Mayr
Christine Mayr
@Mayr_Christine
Something went wrong.
Something went wrong.