Maksim Kuznetsov
@Max__Kuznetsov
Research Scientist at @InSilicoMeds
Anda mungkin suka
I am excited to share that our BindGPT paper won the best poster award at @RealAAAI #AAAI2025! Congratulations to the team! Work led by @artemZholus!

What's the foundational model for generative chemistry? Our work, BindGPT, is a good candidate, and it will be presented at #AAAI2025 today! We built a simple transformer language model that beats diffusion models by just generating 3D molecules as text! Led by @artemZholus 1/n

BindGPT is going to be presented at AAAI 25 today. Work led by my PhD student @artemZholus in collaboration with @InSilicoMeds Don’t miss 10/10 bindgpt.github.io arxiv.org/abs/2406.03686 huggingface.co/insilicomedici…
Excited to present our new paper nach0 from @InSilicoMeds, in collaboration with @nvidia, published in @ChemicalScience: 📄 Paper: pubs.rsc.org/doi/d4sc00966e 💻 Code: github.com/insilicomedici… 🤗 Try it now on @huggingface: huggingface.co/insilicomedici…
Our new paper from @InSilicoMeds on neural conformation generation at @JCIM_JCTC “COSMIC: Molecular Conformation Space Modeling in Internal Coordinates with an Adversarial Framework” Paper: pubs.acs.org/doi/full/10.10… Code: github.com/insilicomedici…

Happy to present our latest results in @InSilicoMeds on molecular graph generation at #AAAI2021! Check out our joint work with @d_polykovskiy “MolGrow: A Graph Normalizing Flow for Hierarchical Molecular Generation” at poster session on 5-Feb, 08:45-10:30 AM & 04:45-06:30 PM PST

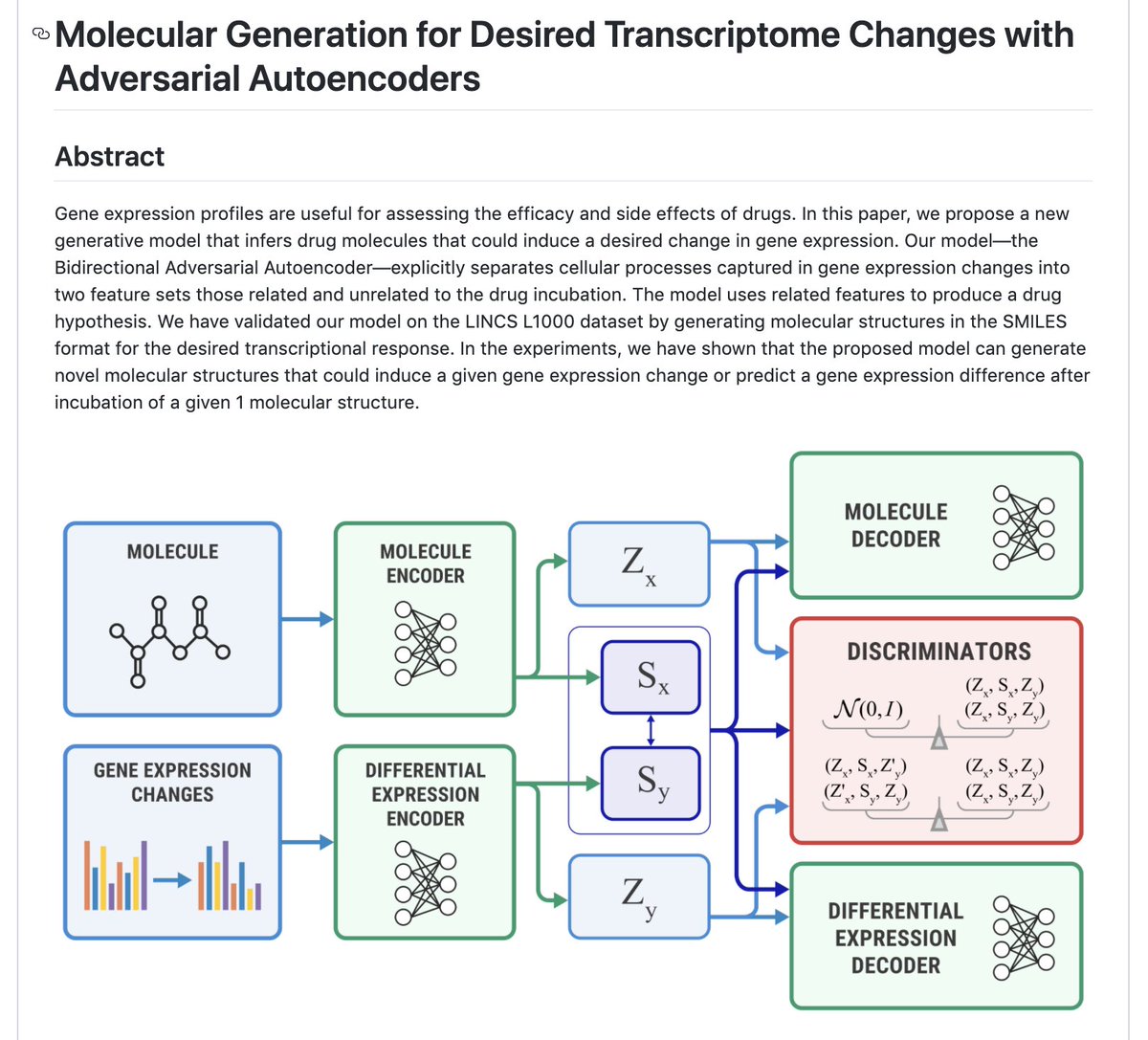
Our new paper with @InSilicoMeds: "Molecular Generation for Desired Transcriptome Changes With AAE". We propose a joint model that can sample molecules for a given transcriptome change and vise versa. Paper: frontiersin.org/articles/10.33… Code: github.com/insilicomedici…

Molecular Generation for Desired Transcriptome Changes with Adversarial Autoencoders. "In this paper, we propose a new generative model that infers drug molecules that could induce a desired change in gene expression" In #pytorch #pytorchLightning github.com/insilicomedici…
Check out our #AISTATS paper from @InSilicoMeds: “Deterministic Decoding for Discrete Data in Variational Autoencoders”. Apparently, for lossless decoding, you need bounded support proposals! arxiv.org/abs/2003.02174


I’m excited to share our new paper at #Neurips2019 with @d_polykovskiy, @alex_zhebrak, and Dmitry Vetrov made in @InSilicoMeds! A Prior of a Googol Gaussians: a Tensor Ring Induced Prior for Generative Models: arxiv.org/abs/1910.13148

Check out a new paper from @InSilicoMeds in @NatureBiotech on the rapid compound hypothesis generation and testing with a powerful combination of variational inference, tensorial methods, and reinforcement learning! paper: nature.com/articles/s4158… code: github.com/insilicomedici…

United States Tren
- 1. GTA 6 6,601 posts
- 2. GTA VI 11.2K posts
- 3. Rockstar 37.9K posts
- 4. #LOUDERTHANEVER 1,465 posts
- 5. Nancy Pelosi 107K posts
- 6. Paul DePodesta 1,374 posts
- 7. Rockies 3,305 posts
- 8. GTA 5 5,729 posts
- 9. Grand Theft Auto VI 29.2K posts
- 10. Ozempic 14K posts
- 11. $TSLA 51.3K posts
- 12. GTA 7 N/A
- 13. RFK Jr 24.9K posts
- 14. Elon Musk 212K posts
- 15. Antonio Brown 2,023 posts
- 16. Marshawn Kneeland 58.3K posts
- 17. Michael Jackson 85.8K posts
- 18. Luke Fickell N/A
- 19. Jonah Hill 1,368 posts
- 20. Subway 45.3K posts
Anda mungkin suka
-
 Nadia Chirkova
Nadia Chirkova
@nadiinchi -
 Bayesian Methods Research Group
Bayesian Methods Research Group
@bayesgroup -
 Polina Kirichenko
Polina Kirichenko
@polkirichenko -
 Artem Sevastopolsky
Artem Sevastopolsky
@ASevastopolsky -
 Kirill Neklyudov
Kirill Neklyudov
@k_neklyudov -
 Ekaterina Lobacheva
Ekaterina Lobacheva
@KateLobacheva -
 Denis Volkhonskiy
Denis Volkhonskiy
@den_volkhonskiy -
 Ivan Sosnovik
Ivan Sosnovik
@isosnovik -
 Evgenii Egorov
Evgenii Egorov
@eeevgen -
 Ilze Amanda Auzina
Ilze Amanda Auzina
@AmandaIlze -
 Talgat Daulbaev
Talgat Daulbaev
@TDaulbaev -
 katushka2ushka
katushka2ushka
@katushka2ushka -
Maxim Panov
@maxim_panov
Something went wrong.
Something went wrong.