
Yu Zhang
@October
Postdoctoral Research Fellow @ Collins Lab, @wyssinstitute, @broadinstitute & @MIT_IMES | AI in drug discovery and synthetic biology
Talvez você curta
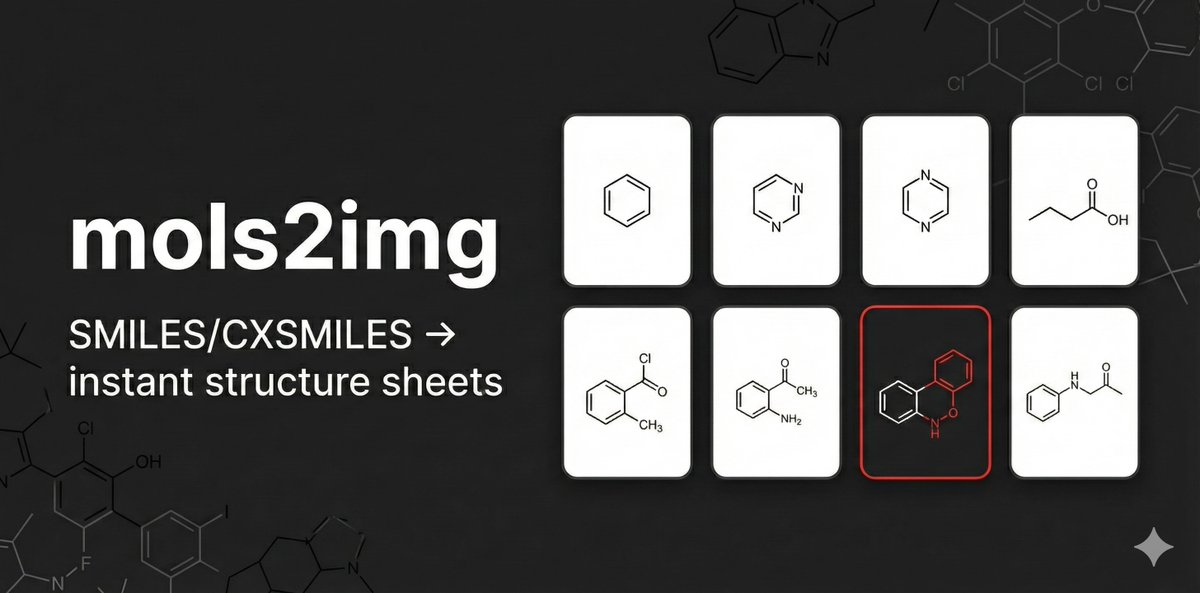
mols2img: paste SMILES/CXSMILES or upload a CSV → instant RDKit structure sheets you can print/save as PDF. Live app: yuzhang-io.github.io/mols2img/
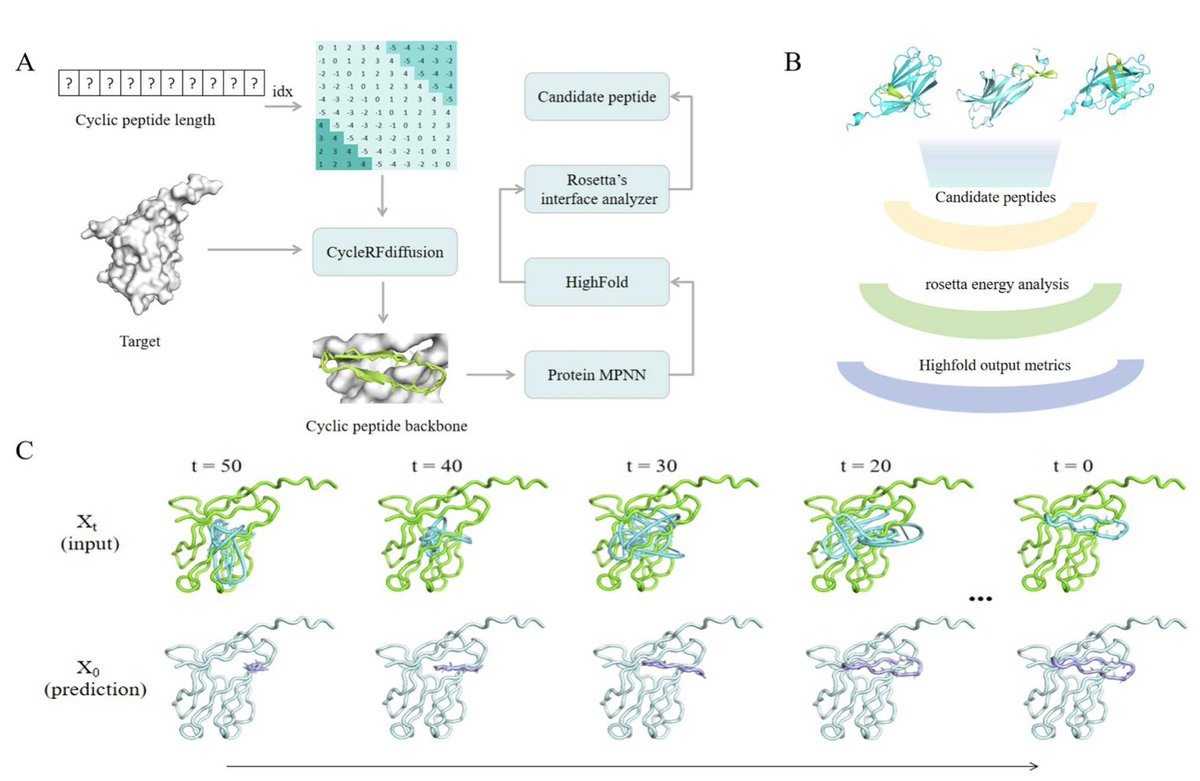
AI-Guided Design of Cyclic Peptide Binders Targeting TREM2 Using CycleRFdiffusion and Experimental Validation 1. A novel study presents an AI-driven pipeline for designing cyclic peptide binders targeting TREM2, a key receptor in neurodegenerative diseases like Alzheimer's. The…
🔥nvMolKit landed today🔥 Morgan Fingerprinting, Tanimoto/Cosine similarity and MMFF geometry optimization and conformer generation on GPU, 10-3000x faster. Screen millions of SMILES before coffee & upsize your QSAR pipelines. 🚀 Which dataset operation will you accelerate…

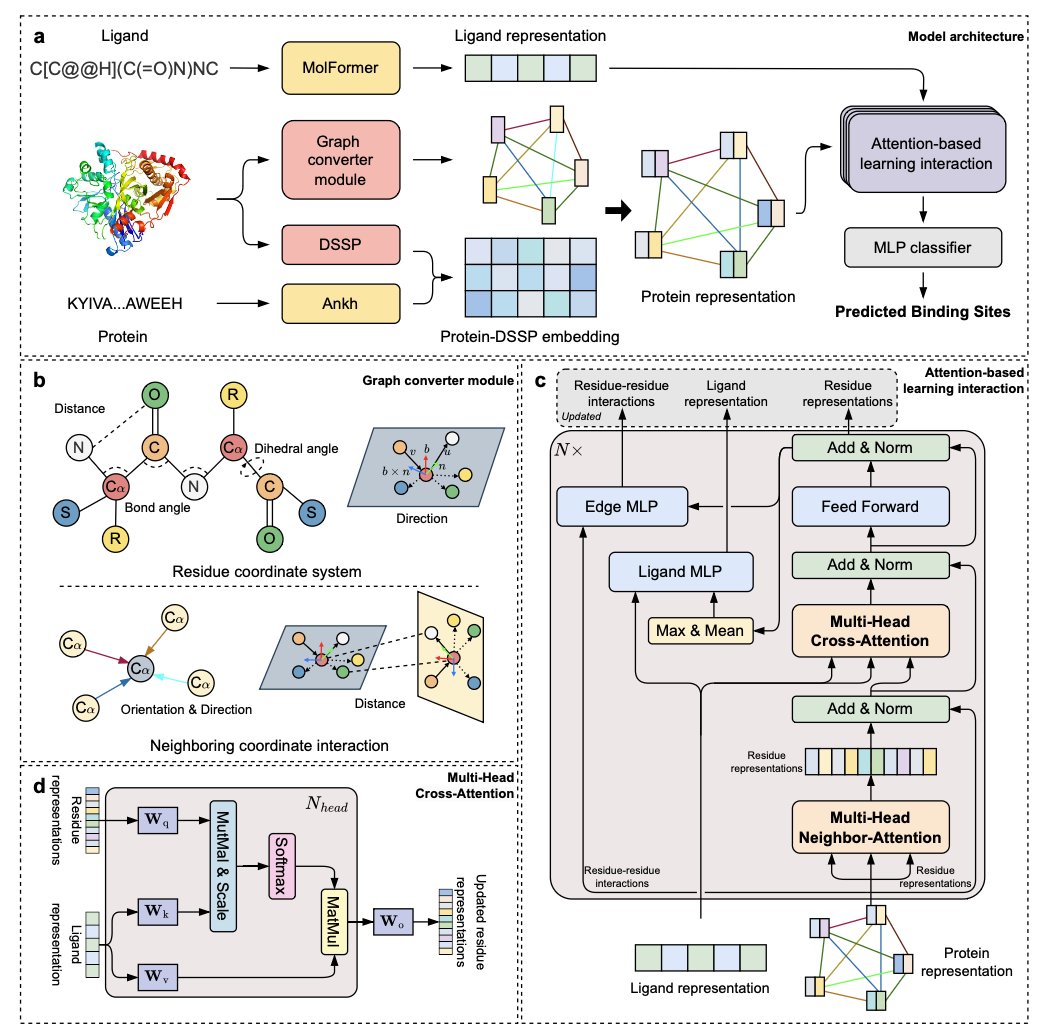
LABind: Identifying Protein Binding Ligand-Aware Sites via Learning Interactions Between Ligand and Protein @NatureComms 1. LABind is a novel structure-based method that predicts protein-ligand binding sites in a ligand-aware manner, utilizing a graph transformer and…
Now online! A generative deep learning approach to de novo antibiotic design dlvr.it/TMWc69
A Generative Deep Learning Approach to De Novo Antibiotic Design @CellCellPress 1. A new generative AI framework has been developed for designing de novo antibiotics, yielding lead compounds with selective antibacterial activity, distinct mechanisms of action, and in vivo…
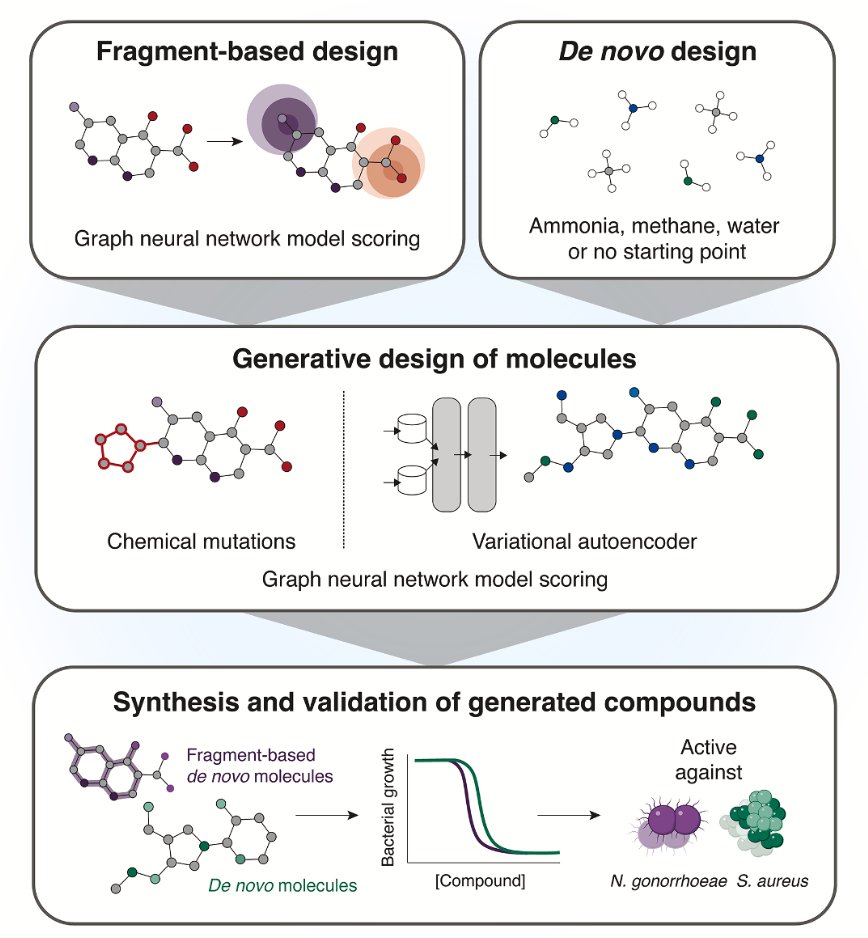
An important application of generative A.I. is facilitating discovery to override antimicrobial resistance with newly designed antibiotics, as demonstrated here for in vivo effectiveness vs S. aureus and N. gonorrhoeae @MIT @broadinstitute @wyssinstitute @MITdeptofBE…
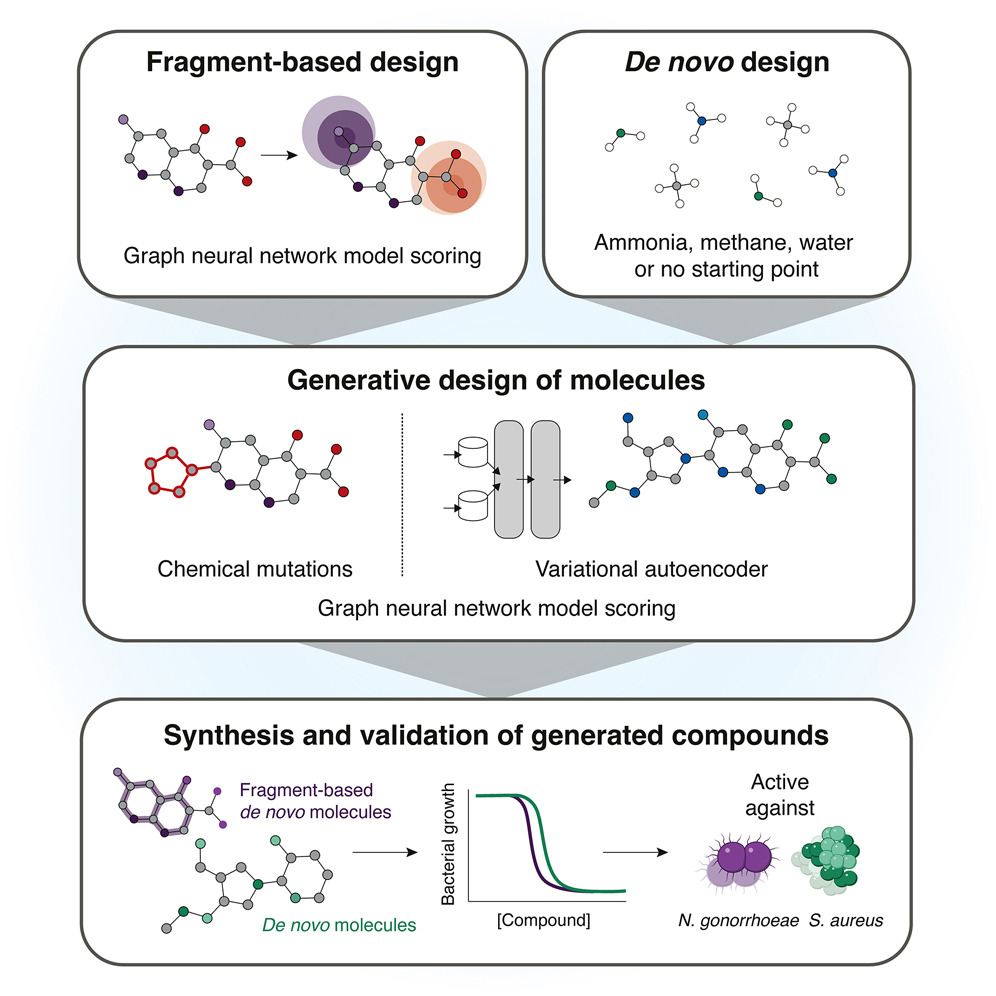
With help from artificial intelligence, MIT researchers have designed novel antibiotics that can combat two hard-to-treat infections: drug-resistant Neisseria gonorrhoeae and multi-drug-resistant Staphylococcus aureus (MRSA). news.mit.edu/2025/using-gen…
MOSAIC lattice light sheet xy projection over 2+ hrs in human retinal pigment epithelial cells of endoplasmic reticulum remodeling (cyan) and transport of vesicles (yellow) containing β4-galactosyltransferase....
New Experimentally Validated De Novo Peptide Design Tool, no structure needed! PepMLM is now fully open-source, try it out on @tamarindbio today. 1/🧵

AI vs. superbugs: who wins? A new Cell study shows how #AI can design new drugs to outsmart deadly bacteria. “We’re building what we think is the most novel and robust pipeline of antibiotics in the world,” says Dr. @AkhilaKosaraju, Phare Bio CEO. 🔗 spectrum.ieee.org/ai-drug-design…
Anthropic literally dropped the smartest one-pager on using AI at work

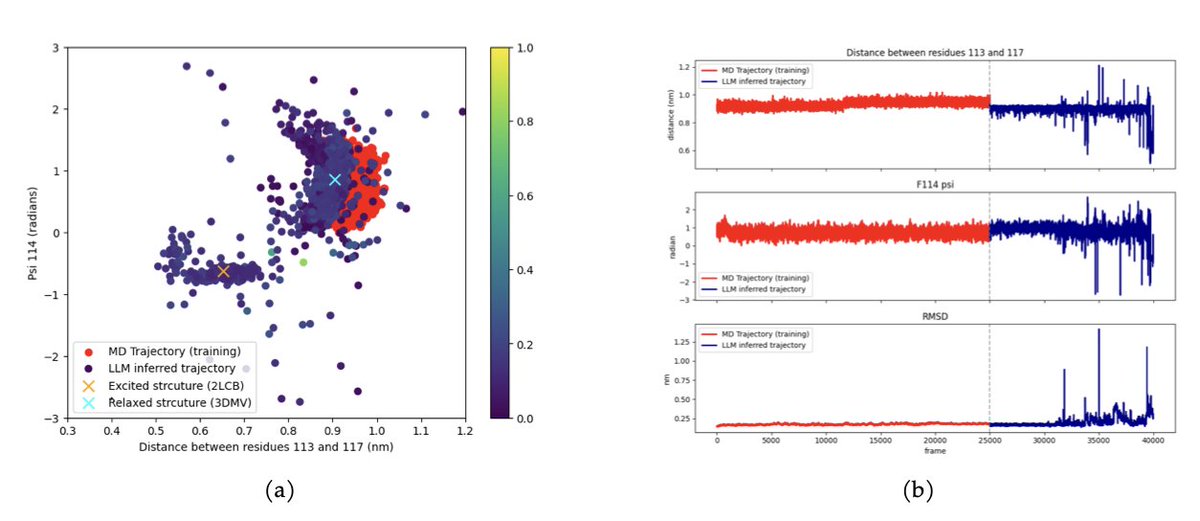
MD-LLM-1: A Large Language Model for Molecular Dynamics 1. Researchers have developed MD-LLM-1, a novel framework that leverages large language models (LLMs) to simulate molecular dynamics. This approach enables the prediction of protein conformational states not seen during…
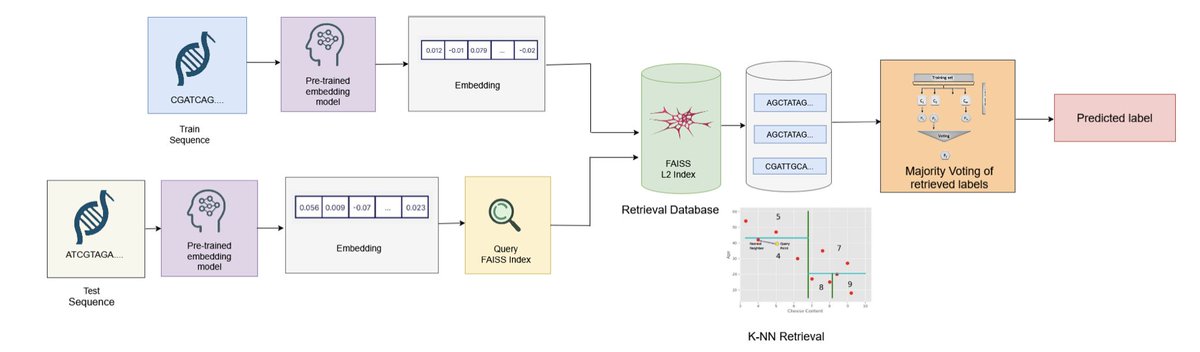
Embedding is (Almost) All You Need: Retrieval-Augmented Inference for Generalizable Genomic Prediction Tasks 1. A new study explores the use of embedding-based pipelines for genomic prediction tasks, challenging the necessity of task-specific fine-tuning of large pre-trained DNA…
Uh oh: deep learning models in genomics lose badly to very simple linear models. Could they have been over-hyped? (new paper by @s_anders_m @const_ae and Wolfgang Huber) nature.com/articles/s4159…
Generative Design of High-Affinity Peptides Using BindCraft 1. A groundbreaking study leverages BindCraft, an AlphaFold-based platform, to design high-affinity peptide ligands directly from protein structures. This approach marks a significant step forward in de novo peptide…

While UK Biobank enabled access to population-based proteomics at scale, most omics studies in disease-focused cohorts still suffer from small sample sizes. The Global Neurodegeneration Proteomics Consortium brought together 35,000 serum, plasma, and CSF samples from…

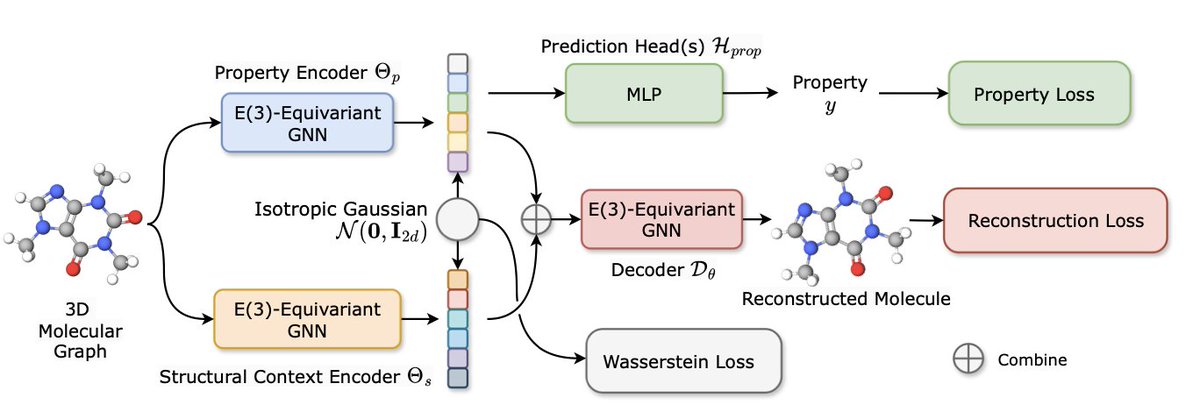
Learning Disentangled Equivariant Representation for Explicitly Controllable 3D Molecule Generation 1. This paper introduces E3WAE, a novel E(3)-equivariant Wasserstein autoencoder designed for explicit control in 3D molecule generation. It factors the latent space into two…
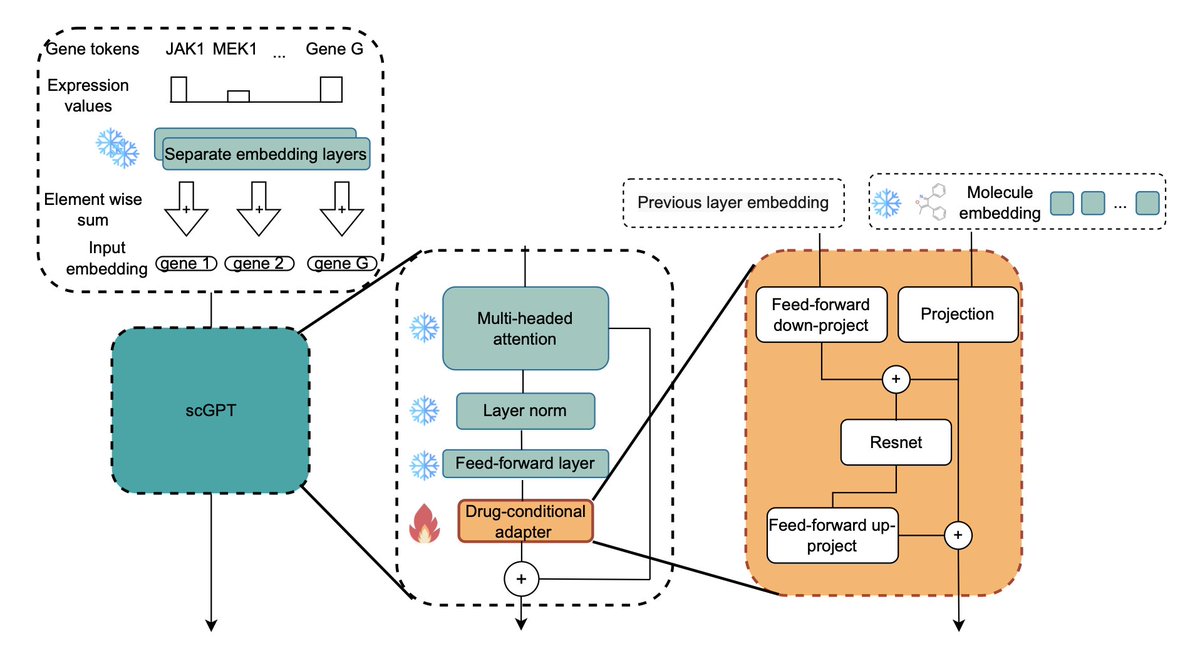
EFFICIENT FINE-TUNING OF SINGLE-CELL FOUNDATION MODELS ENABLES ZERO-SHOT MOLECULAR PERTURBATION PREDICTION @genentech 1. A breakthrough for drug discovery: the study introduces scDCA, a novel drug-conditional adapter, enabling single-cell foundation models (FMs) to predict…
Generative AI for Drug Discovery: A GPT-2 and LSTM Based Models for Designing EGFR Inhibitors • This study explores the use of GPT-2 and LSTM architectures to generate novel inhibitors targeting the Epidermal Growth Factor Receptor (EGFR), a key therapeutic target in cancer.…
United States Tendências
- 1. Dabo N/A
- 2. Blades Brown N/A
- 3. Pete Golding N/A
- 4. Royce Keys N/A
- 5. Wendy N/A
- 6. #ZuffaBoxing01 N/A
- 7. Notre Dame N/A
- 8. #LightningStrikes N/A
- 9. Jassi N/A
- 10. Rivers N/A
- 11. Lobo N/A
- 12. Ryan Wedding N/A
- 13. Deion N/A
- 14. Antarctica N/A
- 15. Stacey N/A
- 16. Saint Louis N/A
- 17. March for Life N/A
- 18. #FursuitFriday N/A
- 19. #ICEOUT N/A
- 20. Jemele N/A
Something went wrong.
Something went wrong.
























































































































