Mohammed Hanzala
@ProteoMatrix
Research Specialist @RockefellerUniv | previously at @UMCG_ERIBA | Mass spectrometry/Proteomics |
Вам может понравиться
The artists & the Art. #Sciart sketched and shaded by the researchers at @IGIBSocial.

Delighted that @teerap16's paper on MAP-MS is out! pubs.acs.org/doi/10.1021/ac… It changes how precursor ions accumulate in Orbitraps to achieve 2× dynamic range without consequences, improving DDA and DIA. It's a free instrument upgrade requiring no special software for Lumos on up!

A LEGO model of Orbitrap Excedion Pro mass spec with a Spectronaut astronaut on top… coolest science souvenir ever! Thank you @ChromSolutions & @biognosys #spectronuat #orbitrap

Time and time again, it's always the same picture. DIA-NN controls FDR correctly as data reliability has been the main goal at DIA-NN's conception back in 2017 and a priority since then. Data from a very nice paper introducing the Thin-diaPASEF concept: doi.org/10.1093/dnares….

We are very excited to officially launch our single cell proteomics service!! Please, check out our website if you want to work with us! sidolilab.org/single-cell

Nielsen et al. track an elusive protein that, over millions of years, has shaped ~45% of the human genome. Despite this profound impact, the protein has evaded detection by conventional methods — making it one of biology’s compelling molecular mysteries. mobilednajournal.biomedcentral.com/articles/10.11…
🧬 The FASEB Mobile DNA conference abstract deadline is fast approaching! 📅 Please register and submit your abstract by May 25th! 🔗 events.faseb.org/event/mobile-d… #FASEB #MobileDNA #Genomics #Transposons #Conference

We’re Hiring: We are looking for a motivated Postdoctoral Researcher for a three-year funded position to contribute to cutting-edge melanoma research as part of the PerMel-AI consortium, funded by EP PerMed. See more details in rug.nl/about-ug/work-… #Proteomics #Bioinformatics
We are hiring! Interested in identifying new therapeutic molecules for obesity and metabolic disorders. Join my lab @csiriiim One project position for biology and one for chemistry @DrShowkatR41235 Apply ⬇️ iiim.res.in/%f0%9f%85%bd%f…
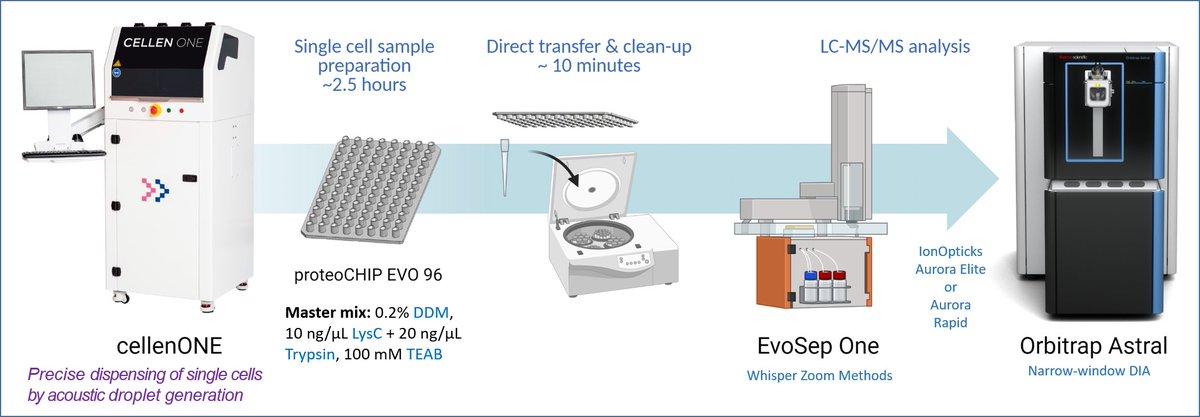
We are excited to share our new Nature Methods paper describing the Chip-Tip workflow for single-cell proteomics identifying >5,000 proteins in single cells, enabling PTM analysis without enrichment and throughput of up to 120 single cell samples per day: rdcu.be/d6qJe
Identification numbers and FDR control of Spectronaut 18 vs DIA-NN 1.9 as benchmarked by Jesper Olsen and colleagues (Nature Methods) nature.com/articles/s4159…

The Mahabaleshwar seminar series set up By Obaid Siddiqi is back! The 43rd version of the student-centric discussion based series covers membranes & organelles. Website: tinyurl.com/5xzkj4y3 Please RT If this area interests you do consider coming to Alibaug

Never in my wildest dreams would I have imagined that the humble DIA tool, I wrote as an intern project at Biognosys, would evolve into what Spectronaut is today. And that fills me with tremendous pride and a feeling of accomplishment.
X-omics training school On July 16 and 17 2024 X-omics will organize an in person workshop “Protein variant detection with proteogenomics data integration”. Please find the program and more information below. #xomics #training #workshop #proteogenomics #dataintegration



Friends, #FragPipe 22 has been released, and it's a big update! diaTracer enables spectrum-centric analysis of diaPASEF data. Skyline integration. Koina server for more deep-learning prediction options. DDA+ mode for ddaPASEF. DIA glycoproteomics and new chemoproteomics workflows

Don't miss our Spectronaut 19 launch seminar on Tuesday morning at #ASMS24. We made a lot of really cool improvements to directDIA, quantification and workflows in general. A shame I can't be there in person this year.
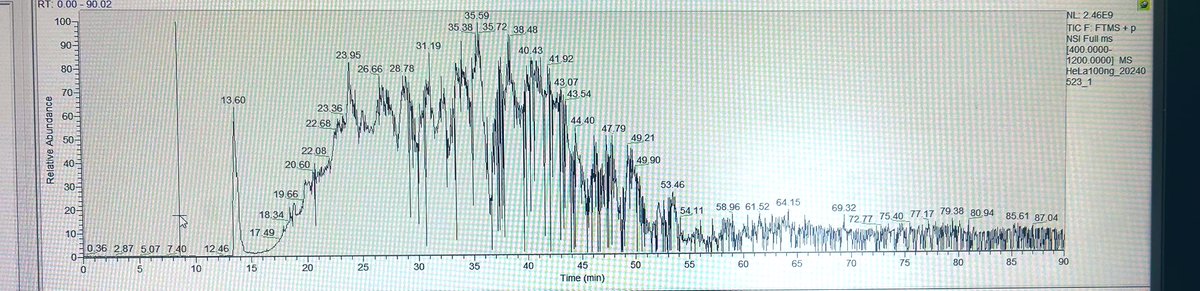
Here’s a snap of TIC from HeLa digest. #TeamMassSpec what could be the reason for this unusual dip in the chromatographic pattern?
Quadrupole out for its yearly scrub-a-dub!

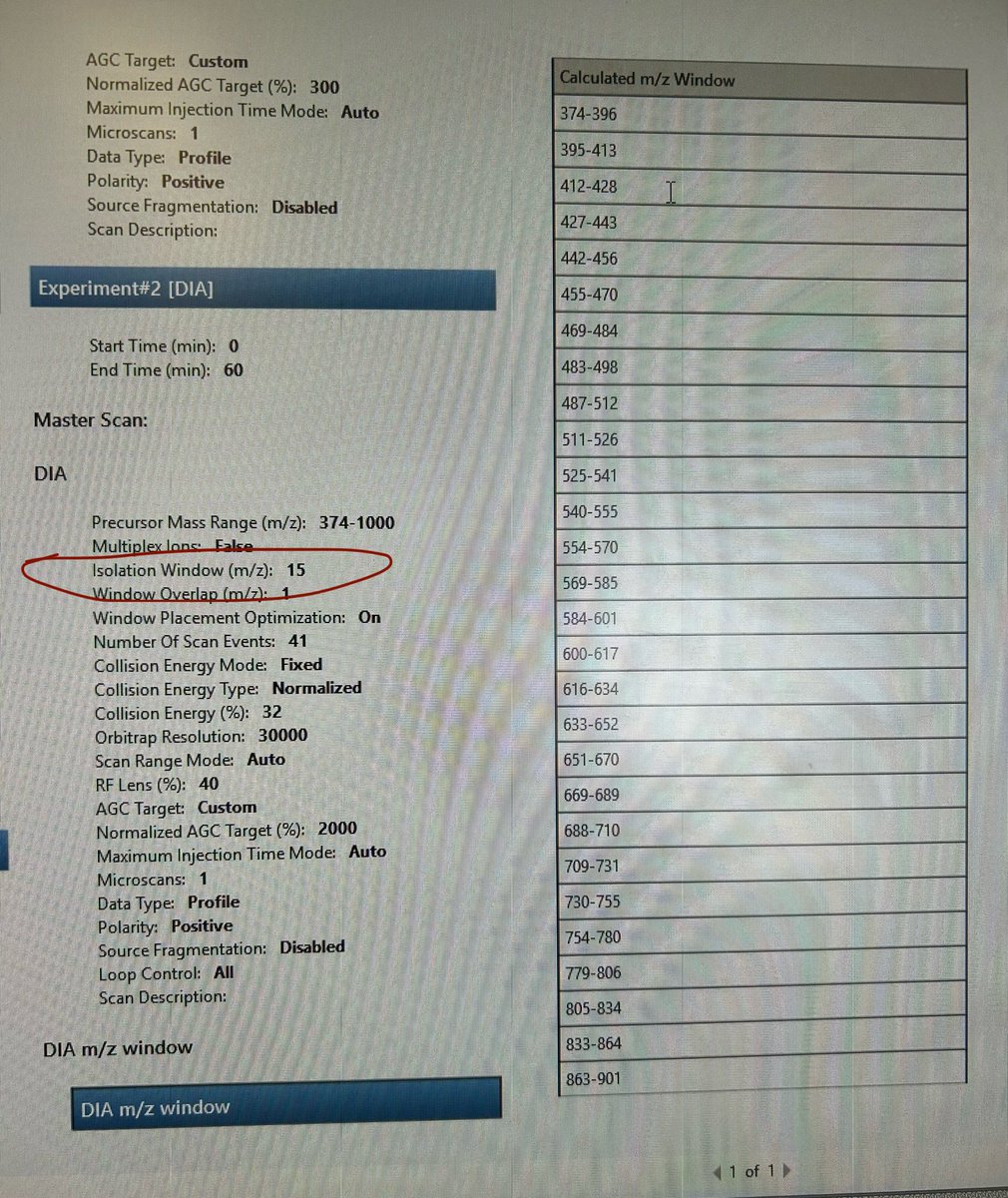
Hi #TeamMassSpec #Proteomics I'm attempting to create a variable m/z window (ranges from 14 to 56 m/z) DIA method on the Exploris 480. Is there a right approach to optimize the Isolation Window parameter from fixed (here it is 15) to variable?
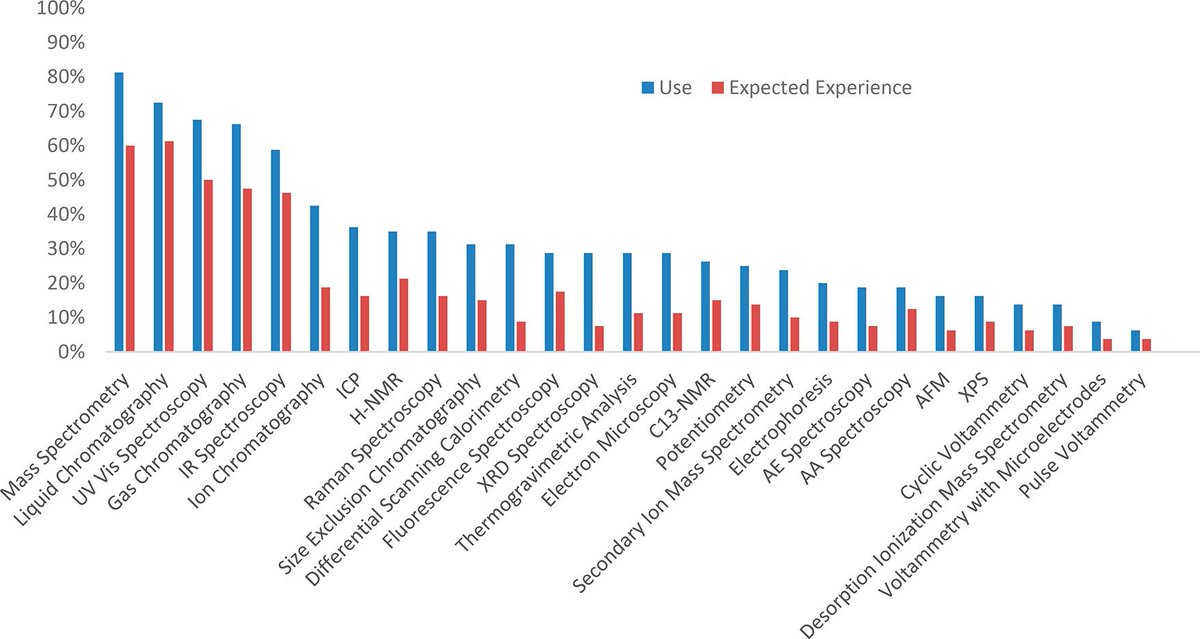
👉Mass Spectrometry is the top instrumentation area where newcomers are expected to have experience. Fresh #JChemEd survey on skills and knowledge needed to best prepare for career in #industry: pubs.acs.org/doi/10.1021/ac… #massspec #CareerAdvice #education
Very nice birthday present for Albert. The first timsOmni trap is currently being delivered to Albert’s lab.



United States Тренды
- 1. Klay 20.3K posts
- 2. #AEWFullGear 70K posts
- 3. McLaren 45.9K posts
- 4. Lando 98.7K posts
- 5. #LasVegasGP 185K posts
- 6. Ja Morant 8,919 posts
- 7. LAFC 15.4K posts
- 8. gambino 2,370 posts
- 9. Hangman 9,878 posts
- 10. Samoa Joe 4,771 posts
- 11. Swerve 6,376 posts
- 12. #Toonami 2,820 posts
- 13. Bryson Barnes N/A
- 14. Verstappen 79.7K posts
- 15. Utah 24K posts
- 16. Kimi 38.6K posts
- 17. Mark Briscoe 4,425 posts
- 18. Benavidez 15.9K posts
- 19. Terry Crews 7,749 posts
- 20. Fresno State 1,004 posts
Something went wrong.
Something went wrong.