Garrett M. Morris
@gmm
(he/him) 🏳️🌈🇪🇺 | @[email protected] | @[email protected] | http://linkedin.com/in/garrettmorris
You might like
We already have some exciting speakers lined up for the 7th @RSC_CICAG / @RSC_BMCS Artificial Intelligence in Chemistry Symposium this September 16-18, 2024, including: • John Jumper, Google DeepMind, UK • Heather Kulik, MIT, USA … rscbmcs.org/events/aichem7/ #AIChem24 1/n
"it's just a tool though, isn't it?" "no it's not, no - it's an alien life form" David Bowie's insights on the internet from 1999 sound exactly like he's talking about AI today
Congratulations to @isakvals on winning the prestigious People’s Poster Prize at the RSC’s 7th "AI in Chemistry" symposium! 🔎Ísak’s research is on developing the AEV-PLIG method within @gmm's group 🔗: stats.ox.ac.uk/news/isak-vals… #AIChem24

Posebusters, work by Martin @Buttenschoen, Garrett Morris (@gmm) & Charlotte Deane that identifies physically implausible molecular binding predictions, was featured by @nathanbenaich and @airstreet Capital in this year's State of AI Report. Full report: stateof.ai/2024-report-la…
Congratulations to @isakvals on winning the RSC’s 7th AI in Chemistry People’s Poster Prize today! :-) Very proud of Ísak and his hard work developing AEV-PLIG in my group: doi.org/10.26434/chemr… Great collab w/ @philbiggin @matthewthwarren & @aniket_magarkar #AIChem24 #AI
Finally, Yael Ziv gave a talk on her recent work MolSnapper, which demonstrates how diffusion models can be effectively conditioned for structure-based drug design. Check out the preprint here:  biorxiv.org/content/10.110…
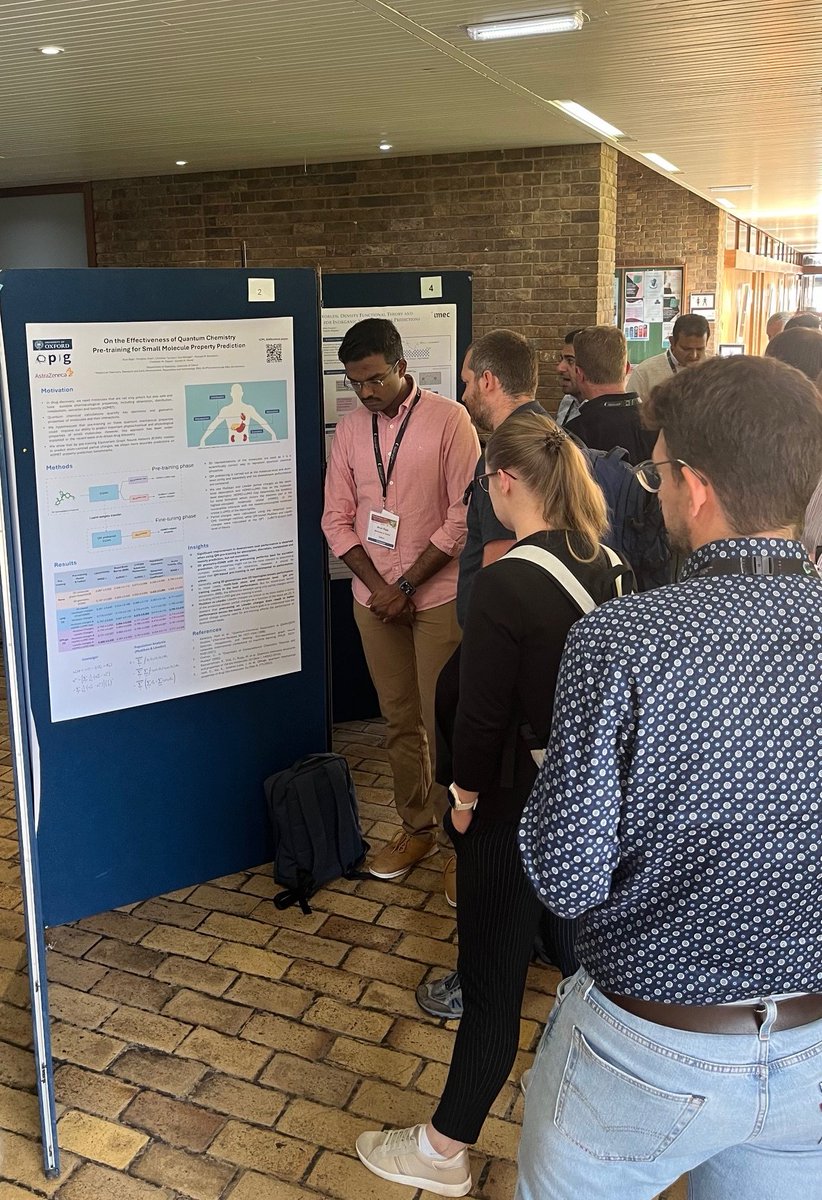
Finally, @arun_raja007 has explored the effectiveness of quantum chemistry pre-training GNNs for pharmacological property prediction openreview.net/pdf?id=ffSLXl6…
@LucyVost tells us how we can improve chemical plausibility in 3D molecule generation via conditional training with distorted molecules (preprint coming soon!)

First the posters! @isakvals is presenting his recent work on AEV-PLIG, a machine learning scoring function for binding affinity prediction chemrxiv.org/engage/chemrxi…

OPIGlets are out in force at #AIChem24, organised by our very own @gmm DPhil students @LucyVost, @arun_raja007 and @isakvals are presenting posters, and Yael is giving a talk about her recent work MolSnapper on Wednesday! Please come and chat to them!
We're delighted to be hosting @dr_greg_landrum on Mon Nov 20th at Comp Chem Kitchen CCK-20 — 5 pm GMT in @OxfordStats 24-29 St Giles', OX1 3LB; register for Zoom. Refreshments at 4:30 pm. compchemkitchen.org/2023/10/24/sav… @CompChemKitchen @philbiggin @coopallographer @fjduarteg @bobbypaton
We are excited - and proud - report that, over the last 9 months, BDB curated and shared103K open-access, machine-readable protein-ligand binding data, melding it with the 74K new binding data curated by ChEMBL, for a total 177K increase in our holdings.

Talk: "John Mayfield: Managing and Searching One Trillion Compounds" presented at UCSF DOCK Meeting. #cheminformatics #Arthor #SmallWorld nextmovesoftware.com/talks/Mayfield…

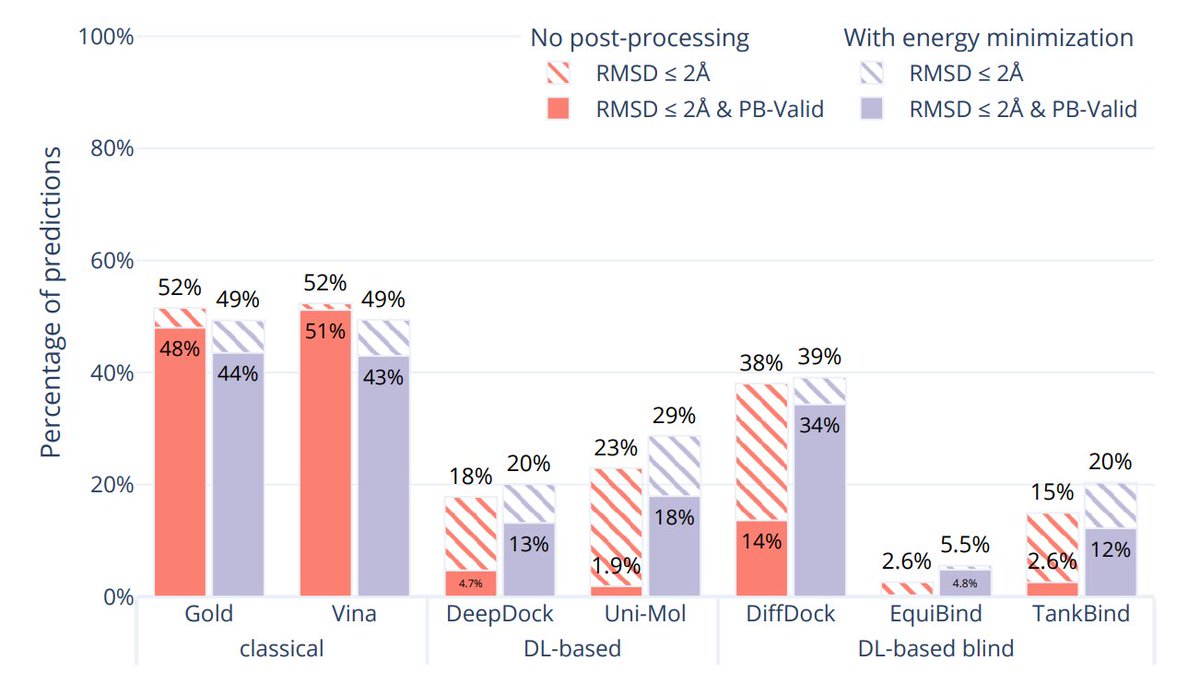
The new PoseBusters paper does some super neat docking experiments! arxiv.org/abs/2308.05777 E.g. how DL methods generalize to different levels of seq similarity. And impact of relaxing DL structures on steric clashes etc. captured in the PoseBusters score.

United States Trends
- 1. Louisville 12.7K posts
- 2. Nuss 5,330 posts
- 3. Miller Moss N/A
- 4. Bama 12.5K posts
- 5. Ty Simpson 3,002 posts
- 6. The ACC 19.2K posts
- 7. #AEWCollision 9,114 posts
- 8. Hawks 16.5K posts
- 9. Lagway 3,351 posts
- 10. Clemson 6,150 posts
- 11. Kentucky 30.2K posts
- 12. Brohm N/A
- 13. Emmett Johnson N/A
- 14. Van Buren 1,233 posts
- 15. Stoops 1,809 posts
- 16. #RollTide 4,886 posts
- 17. #RockHall2025 4,558 posts
- 18. Wake Forest 2,017 posts
- 19. Watford 3,065 posts
- 20. #OrtizLubin 1,969 posts
You might like
Something went wrong.
Something went wrong.