Danny IncaRNAto
@incaRNAtolab
Associate Professor @univgroningen @ResearchGbb | Chief R&D Officer @serna_bio | #RNA structure ensemble dynamics | 🦋 http://incarnatolab.bsky.social
You might like
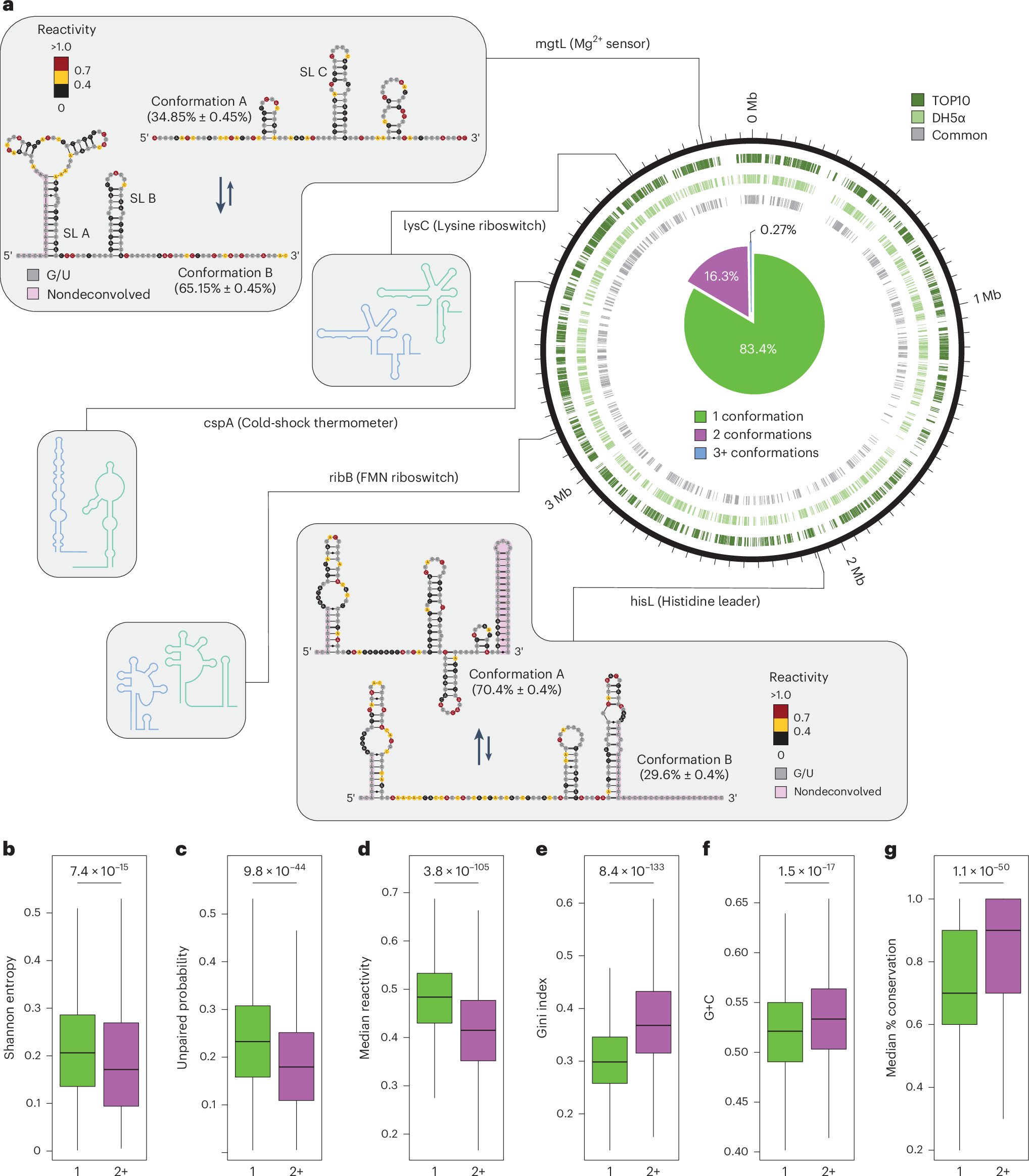
I am so incredibly excited to share our latest work, on the exploration of #RNA secondary structure ensembles and discovery of RNA regulatory structural switches in bacteria and human cells, out today in @NatureBiotech: nature.com/articles/s4158…. A short tread! (1/n)
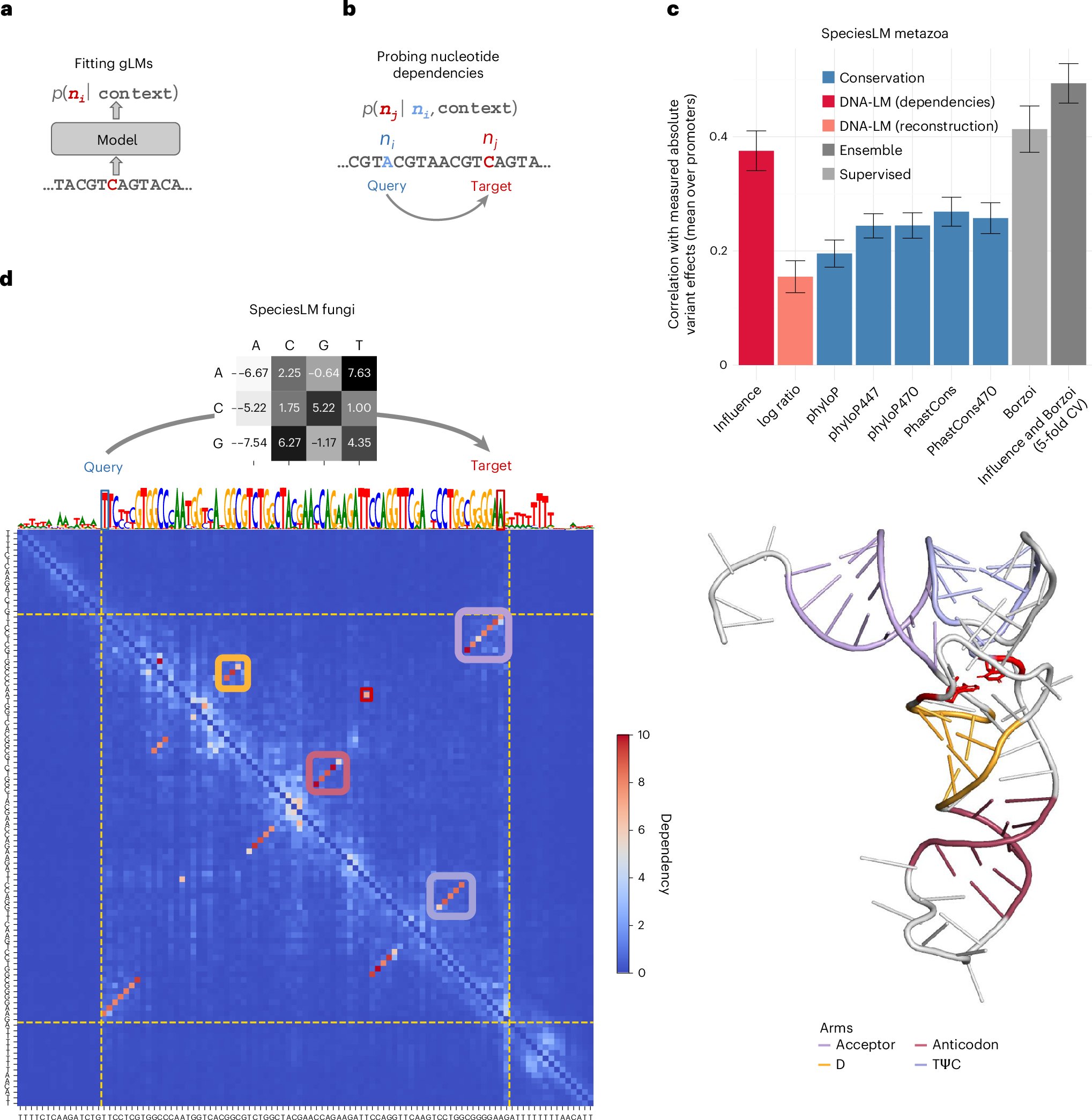
So excited to see this out in @NatureGenet! An amazing collaboration with the @gagneurlab I am happy I was (a small) part of... nucleotide dependencies can capture regulatory elements, including #RNA structures! Congrats to the whole team! Check it out: nature.com/articles/s4158…
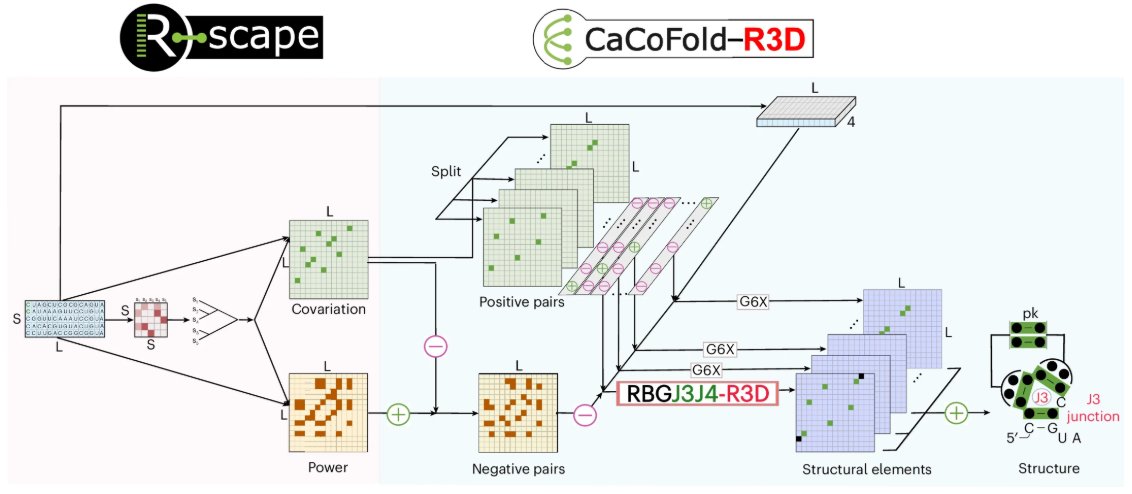
CaCoFold-R3D: a probabilistic model that predicts RNA 3D motifs and secondary structure using evolutionary information. nature.com/articles/s4159…
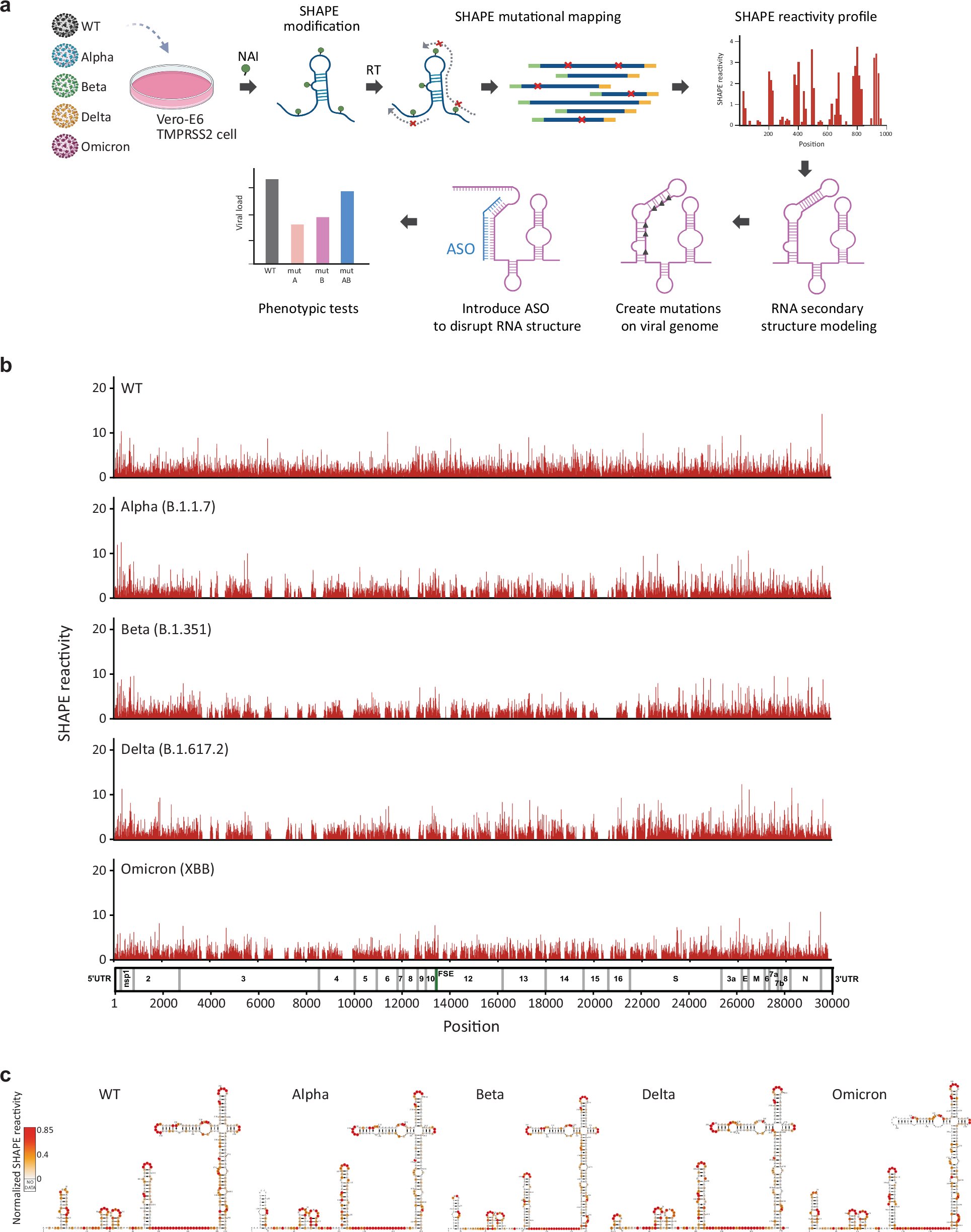
Happy to share that we identified a conserved 17kb long-range RNA-RNA interaction in SARS-CoV-2 that impacts virus fitness. Congratulations to all the authors! nature.com/articles/s4146…
Termal: a fast and interactive terminal-based viewer for multiple sequence alignments. #MultipleSequenceAlignments #TerminalViewer #interactiveViewer #Bioinformatics @BioinfoAdv academic.oup.com/bioinformatics…

A great opportunity to work with one of the best. If you love #RNA and plants, this is for you!
Postdoctoral Researcher (Ding-Dean Group) | John Innes Centre jic.ac.uk/vacancies/post…
Excited to have this work finally published on @NatureComms. With the Trcek lab @JohnsHopkins, @silvirouskin and @ThirumalaiArmy labs, we show that RNA structure impedes intermolecular base pairing in germ granule. nature.com/articles/s4146…
I'm excited to share our new preprint on LagTag, a method that recovers both past and present chromatin states from the same mammalian cells. biorxiv.org/content/10.110…

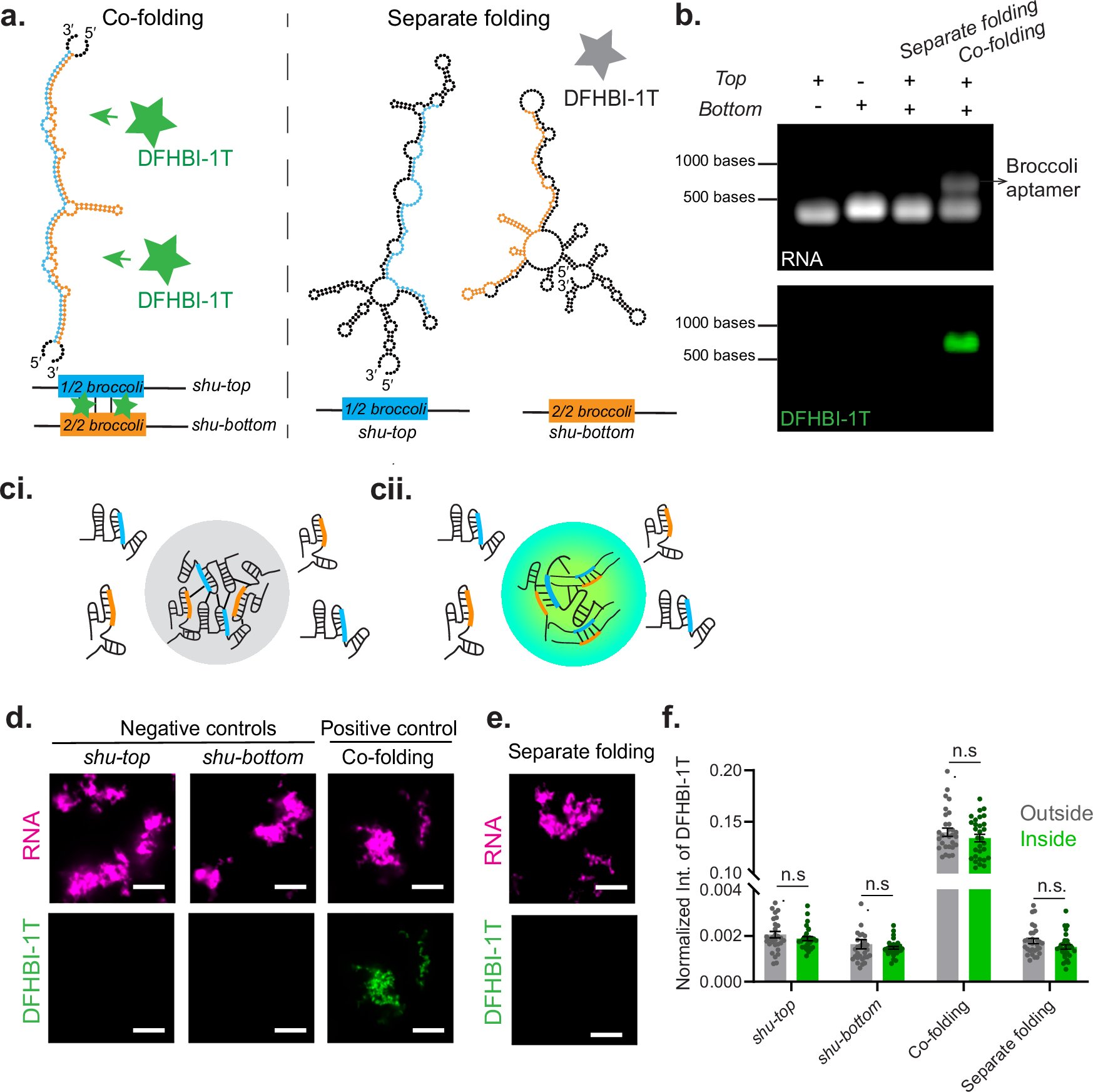
New online! Structural insights into higher-order natural RNA-only multimers bit.ly/4m6opSt

Excited to share our latest work with the bullock lab! We looked at how diverse mRNAs get selected for subcellular localization and it turns out that a single protein can recognize different RNA elements using shared features that weren’t apparent before. biorxiv.org/content/10.110…
United States Trends
- 1. #GMMTV2026 588K posts
- 2. MILKLOVE BORN TO SHINE 86.5K posts
- 3. #WWERaw 77K posts
- 4. Finch 14.6K posts
- 5. Purdy 28.4K posts
- 6. AI Alert 8,053 posts
- 7. TOP CALL 9,278 posts
- 8. Bryce 21.3K posts
- 9. Keegan Murray 1,538 posts
- 10. Alan Dershowitz 2,735 posts
- 11. Barcelona 136K posts
- 12. Timberwolves 3,901 posts
- 13. Check Analyze 2,418 posts
- 14. Token Signal 8,589 posts
- 15. Gonzaga 4,118 posts
- 16. Panthers 37.8K posts
- 17. Dialyn 7,738 posts
- 18. Enemy of the State 2,594 posts
- 19. Market Focus 4,702 posts
- 20. Niners 5,928 posts
You might like
-
 RNA
RNA
@RNAJournal -
 Kwok Lab
Kwok Lab
@kitkwok6 -
 Woodson Lab
Woodson Lab
@woodson_lab -
 The RNA Society
The RNA Society
@RNASociety -
 RNAcentral
RNAcentral
@RNAcentral -
 UMich RNA Center 〽️🧬
UMich RNA Center 〽️🧬
@umichrna -
 Nils Walter Lab
Nils Walter Lab
@NilsWalterLab -
 Jinwei Zhang Lab
Jinwei Zhang Lab
@jinweizhang -
 Jr RNA Scientists
Jr RNA Scientists
@jrRNAscientists -
 Das Lab
Das Lab
@RDasLab -
 Yiliang Ding
Yiliang Ding
@YiliangDing -
 The RNA Institute
The RNA Institute
@TheRNAInstitute -
 Igor Ulitsky
Igor Ulitsky
@IgorUlitsky -
 Robert Spitale
Robert Spitale
@SpitaleLab
Something went wrong.
Something went wrong.