Jason Yim
@json_yim
PhD student @MIT_CSAIL. Generative models, protein design. 🦋 Bluesky handle: https://bsky.app/profile/jyim.bsky.social On X until the exodus is complete.
قد يعجبك
Combining discrete and continuous data is an important capability for generative models. To address this for protein design, we introduce Multiflow, a generative model for structure and sequence generation. Preprint: arxiv.org/abs/2402.04997 Code: github.com/jasonkyuyim/mu… 1/8
We introduce a new ''rule'' for understanding diffusion models: Selective Underfitting. It explains: 🚨 How diffusion models generalize beyond training data 🚨 Why popular training recipes (e.g., DiT, REPA) are effective and scale well Co-led with @kiwhansong0! (1/n)

It's odd the performance is significantly worse than the AR base model? Starting with a much powerful AR model, dropping the performance just enough to beat all other diffusion LLMs, and then saying it's better than them is weird...

More on RND1 models: Blog: radicalnumerics.ai/blog/rnd1 Code: github.com/RadicalNumeric… Report: radicalnumerics.ai/assets/rnd1_re… Weights: huggingface.co/radicalnumeric…

New work: “GLASS Flows: Transition Sampling for Alignment of Flow and Diffusion Models”. GLASS generates images by sampling stochastic Markov transitions with ODEs - allowing us to boost text-image alignment for large-scale models at inference time! arxiv.org/pdf/2509.25170 [1/7]
We've cleaned up the story big time on flow maps. Check out @nmboffi's slick repo implementing all the many ways to go about them, and stay tuned for a bigger release 🤠 arxiv.org/pdf/2505.18825 flow-maps.github.io
Consistency models, CTMs, shortcut models, align your flow, mean flow... What's the connection, and how should you learn them in practice? We show they're all different sides of the same coin connected by one central object: the flow map. arxiv.org/abs/2505.18825 🧵(1/n)
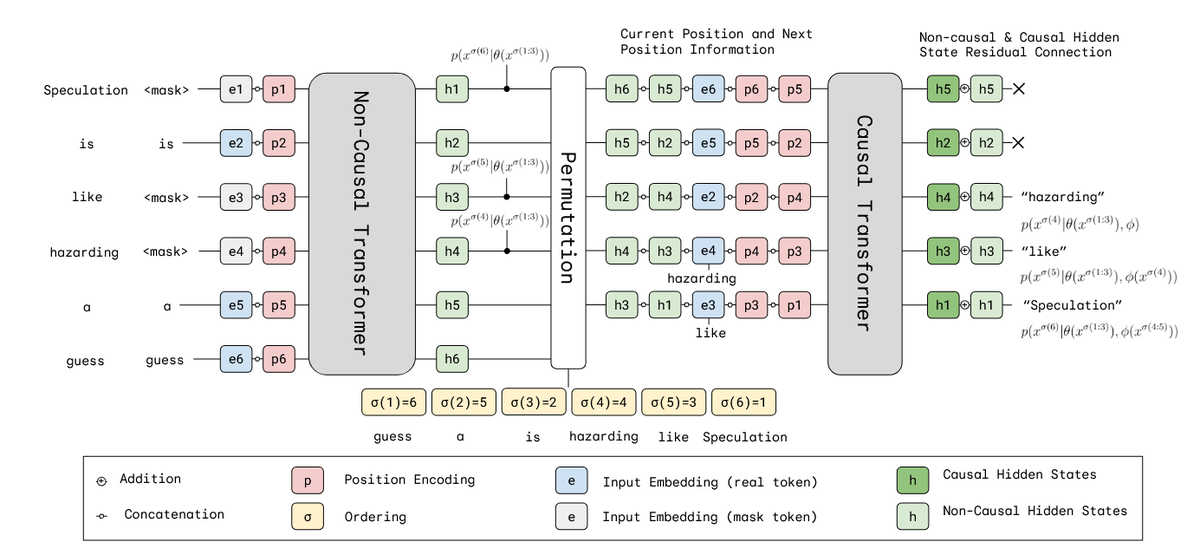
Very excited to share our preprint: Self-Speculative Masked Diffusions We speed up sampling of masked diffusion models by ~2x by using speculative sampling and a hybrid non-causal / causal transformer arxiv.org/abs/2510.03929 w/ @ValentinDeBort1 @thjashin @ArnaudDoucet1
🎉Personal update: I'm thrilled to announce that I'm joining Imperial College London @imperialcollege as an Assistant Professor of Computing @ICComputing starting January 2026. My future lab and I will continue to work on building better Generative Models 🤖, the hardest…
We've open sourced Adjoint Sampling! It's part of a bundled release showcasing FAIR's research and open source commitment to AI for science. github.com/facebookresear… x.com/AIatMeta/statu…
Announcing the newest releases from Meta FAIR. We’re releasing new groundbreaking models, benchmarks, and datasets that will transform the way researchers approach molecular property prediction, language processing, and neuroscience. 1️⃣ Open Molecules 2025 (OMol25): A dataset…
#FPIworkshop best paper award goes to @peholderrieth @msalbergo and Tommi Jaakkola. Congrats and great talk Peter!


I won't be at ICLR 🥲 but you can talk to these other cool people at my poster, Thursday 3-5:30 PM in Hall 3+2B #10!
Excited to share my #ICLR2025 paper, with JC Hütter and friends! Genetic perturbation screens allow biologists to manipulate and measure the genes in cells = discover causal relationships! BUT they are expensive to run, expensive to interpret. ... We use LLMs to help!

Had fun exploring guidance for backbone designability within this latent framework, excited to chat more about guidance with experimental data @gembioworkshop ICLR

I'll be at the ICLR @gembioworkshop workshop presenting latent and structure diffusion for protein backbone generation. Come by to talk all things latent for biology. openreview.net/forum?id=Ek7Hs… arxiv.org/abs/2504.09374
I'll be at the ICLR @gembioworkshop workshop presenting latent and structure diffusion for protein backbone generation. Come by to talk all things latent for biology. openreview.net/forum?id=Ek7Hs… arxiv.org/abs/2504.09374
I'll be at ICLR. Come check out our generative modeling work! Reach out if you want to chat. Proteina: x.com/karsten_kreis/… Protcomposer: x.com/HannesStaerk/s… Generator matching: x.com/peholderrieth/…
New paper out! We introduce “Generator Matching” (GM), a method to build GenAI models for any data type (incl. multimodal) with any Markov process. GM unifies a range of state-of-the-art models and enables new designs of generative models. arxiv.org/abs/2410.20587 (1/5)

RFdiffusion => generative binder design. RFdiffusion2 => generative enzyme design. It's rare to find scientists with deep knowledge in chemistry, machine learning, and software engineering like Woody. The complexity of enzymes matches the complexity of his skills. Check out RFD2
New enzymes can unlock chemistry we never had access to before. Here, we introduce RFdiffusion2 (RFD2), a generative model that makes significant strides in de novo enzyme design. Preprint: biorxiv.org/content/10.110… Code: coming soon Animation credit: x.com/ichaydon (1/n)
New enzymes can unlock chemistry we never had access to before. Here, we introduce RFdiffusion2 (RFD2), a generative model that makes significant strides in de novo enzyme design. Preprint: biorxiv.org/content/10.110… Code: coming soon Animation credit: x.com/ichaydon (1/n)
Excited to share our preprint “BoltzDesign1: Inverting All-Atom Structure Prediction Model for Generalized Biomolecular Binder Design” — a collaboration with @MartinPacesa, @ZhidianZ , Bruno E. Correia, and @sokrypton. 🧬 Code will be released in a couple weeks
Protein dynamics was the first research to enchant me >10yrs ago, but I left in PhD bc I couldnt find big expt data to evaluate models. Today w @ginaelnesr, I'm thrilled to share the big dynamics data I've been dreaming of, and the mdl we trained: Dyna-1. rb.gy/de5axp

Combining prediction, generation, and modalities (sequence, structure, nucleic acids, small molecules, proteins) is the future. Congrats to the authors! Looking forward to the technical report.
Introducing All-atom Diffusion Transformers — towards Foundation Models for generative chemistry, from my internship with the FAIR Chemistry team @OpenCatalyst @AIatMeta There are a couple ML ideas which I think are new and exciting in here 👇

Awesome work by Hannnes and Bowen towards improved control of protein structure generation with MultiFlow!
New paper (and #ICLR2025 Oral :)): ProtComposer: Compositional Protein Structure Generation with 3D Ellipsoids arxiv.org/abs/2503.05025 Condition on your 3D layout (of ellipsoids) to generate proteins like this or to get better designability/diversity/novelty tradeoffs. 1/6
I really enjoyed seeing how protein generation models scale with more data and weights. Congrats to Nvidia and the core contributors for this amazing work!
📢📢 "Proteina: Scaling Flow-based Protein Structure Generative Models" #ICLR2025 (Oral Presentation) 🔥 Project page: research.nvidia.com/labs/genair/pr… 📜 Paper: arxiv.org/abs/2503.00710 🛠️ Code and weights: github.com/NVIDIA-Digital… 🧵Details in thread... (1/n)
United States الاتجاهات
- 1. Jets 104K posts
- 2. James Franklin 37.6K posts
- 3. Drake Maye 11.1K posts
- 4. Justin Fields 20.6K posts
- 5. Penn State 52.2K posts
- 6. Broncos 44.3K posts
- 7. Aaron Glenn 8,657 posts
- 8. Puka 7,212 posts
- 9. Derrick Henry 2,334 posts
- 10. George Pickens 4,014 posts
- 11. Steelers 41.6K posts
- 12. Cooper Rush 1,830 posts
- 13. #RavensFlock 1,654 posts
- 14. Saints 46.6K posts
- 15. Cam Little N/A
- 16. Boutte 2,546 posts
- 17. Tyler Warren 2,020 posts
- 18. Sean Payton 3,673 posts
- 19. London 206K posts
- 20. #DallasCowboys 2,088 posts
قد يعجبك
-
 Generate:Biomedicines
Generate:Biomedicines
@generate_biomed -
 Namrata Anand
Namrata Anand
@namrata_anand2 -
 Brian Hie
Brian Hie
@BrianHie -
 David Juergens
David Juergens
@DaveJuergens -
 Alex Chu
Alex Chu
@alexechu_ -
 Gabriele Corso
Gabriele Corso
@GabriCorso -
 chris bahl
chris bahl
@bahl_lab -
 Tom Sercu
Tom Sercu
@TomSercu -
 Jue Wang
Jue Wang
@jueseph -
 Roshan Rao
Roshan Rao
@proteinrosh -
 Helen Eisenach O'Brien
Helen Eisenach O'Brien
@HelenEisenach -
 Amir Motmaen
Amir Motmaen
@AMotmaen -
 Zuobai Zhang
Zuobai Zhang
@Oxer22 -
 Sam Tipps
Sam Tipps
@SamTipps -
 Soojung Yang
Soojung Yang
@SoojungYang2
Something went wrong.
Something went wrong.