Maxime Rousseaux
@maximewc
Assistant Professor and CRC studying protein mislocalization, neurodegeneration and functional genomics @uOttawaBMRI @OISB_ISBO. Co-director @GEMb_uOttawa
You might like
🚨At long last: our lab's first manuscript on #ALS/#FTD, describing stress-responsive #TDP43 SUMOylation as a neuroprotective mechanism. Very proud of the collective work of many Rousseaux lab members, spearheaded by @TerryRSuk. Please see his tweetorial below. Feedback welcome!
Great collaborating with the Russell lab on this paper. Tireless efforts from first author, Truc (Cloe) Losier, particularly in optimizing ideal screening conditions - protocol paper incoming! Nice work @OISB_IBSO @uOttawaMed
Truc Losier, Ryan Russell et al. @uOttawa identify distinct kinase signaling pathways that differentially regulate bulk and selective #autophagy. Using kinome-wide #CRISPR screening, they reveal both shared and pathway-specific regulators hubs.la/Q03DSv6d0

SUMO to the rescue! Three new studies show that tagging TDP-43 with SUMO2/3 keeps it soluble under stress, preventing the toxic aggregates that mark ALS and FTD. @uOttawa, @UNIMORE_Univ, @goetheuni ow.ly/ROwi50VQk61
'A stress-dependent #TDP43 SUMOylation program preserves neuronal function' Terry R. Suk @TerryRSuk Caroline E. Part, Jenny L. Zhang, Trina T. Nguyen...Maxime W. C. Rousseaux @maximewc @uOttawaBMRI @uOttawa #ALS …arneurodegeneration.biomedcentral.com/articles/10.11…

Thrilled to see our work published in its final form @MolNeuro . Outstanding work by @TerryRSuk and colleagues. Please check-out the tweetorial below. As always, happy to share reagents (plasmids are being put on @Addgene), mice and other data; just reach out!
We’re happy to share that our labs first #ALS and #FTD study “A stress-dependent TDP-43 SUMOylation program preserves neuronal function” is now online @MolNeuro! doi.org/10.1186/s13024…
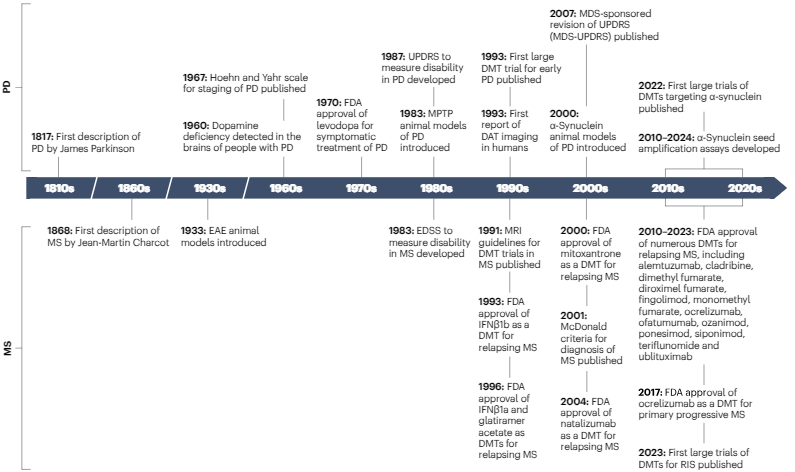
Where are we for disease modifying therapies and Parkinson's? Why not take some lessons from multiple sclerosis? Fantastic paper by Lorraine Kalia and colleagues in @Nature Reviews Neurology. Can you say MRI? Key Points: - Guess what? ~18 disease-modifying therapies or DMTs for…

I am once again posting our open 3-5 years postdoctoral position on proteostasis and aging, emphasizing that we are in beautiful, human rights-friendly and politically-stable New Brunswick, Canada. Bernie would be a regular guy here. Come join our team! postdocjobs.com/posting/728638…
Cold-emailing potential postdoc advisors is a hugely successful strategy! Here is the exact e-mail I sent back in the day (& it worked) - feel free to use as a template: docs.google.com/document/d/1Ob…
There is much we can learn from each other. Read the ideas on DMT discovery that came from @UofTNeurology bringing experts in #Parkinsons and #multiplesclerosis together in #Toronto (published with @SpringerNature in @NatRevNeurol) rdcu.be/dWf2U
We are so proud of our team, who worked relentlessly on this project. Read about Using AI to improve the diagnosis of rare genetic disorders here: bcm.edu/news/using-ai-…
How to pick a problem. Today in @CellCellPress. 1/53 cell.com/cell/fulltext/…
A new Parkinson’s gene has been discovered, RAB32. This gene is possibly involved in the same pathway as LRRK2. Great discovery by Matt Farrer and his team (who initially discovered this gene as a candidate back in 2017). sciencedirect.com/science/articl…
Reminder to apply! Come do great science in beautiful #Ottawa!
🧠🚨Our lab is hiring! Please spread the word. Multiple exciting projects in the lab including a fully funded postdoctoral fellowship/research associate position. @uOttawa @uOttawaBMRI @ALSCanada @CAN_ACN

New paper from the lab: Genetic and pharmacological reduction of CDK14 mitigates synucleinopathy. Congratulations to to all of those involved and thanks to @ASAP_Research and @CIHR_IRSC for the support. @uOttawaBMRI @uOttawaMed nature.com/articles/s4141…
We are excited to share this comparative dataset of selective and bulk #autophagy regulators. Thanks to @maximewc for the collaboration, which created a new research focus in the lab. Comments or questions welcome.
Happy to share the latest from the lab. A fun collaboration with @Russell_BioLab looking at stress-specific regulators of #autophagy using tandem pooled kinome screens. Feedback most welcome. biorxiv.org/content/10.110…
Happy to share the latest from the lab. A fun collaboration with @Russell_BioLab looking at stress-specific regulators of #autophagy using tandem pooled kinome screens. Feedback most welcome. biorxiv.org/content/10.110…
Research Associate/Postdoctoral Fellow in Rousseaux lab at uOttawa @maximewc We are actively recruiting a Postdoctoral Fellow/Research Associate to study mechanisms underlying ALS and FTD. jobrxiv.org/job/university… #als #animal_behavior #biochemistry #...
Unpopular opinion coming up. I screen a bunch of preprints for BioRxiv, have done for a decade. I think these are fairly representative of what is published generally - e.g. similar in quality, etc., even though only ~20% of papers are preprinted. /1
New paper from the lab from the talented @HaleyMGeertsma detailing aSyn topography in the mouse brain. Implications for #Parkinsonsdisease. Thanks to @ASAP_Research for the support and to @MXHend for the fun collaboration. See below for tweetorial
Check it out! 🧐 We used our recently developed Snca-NLS mouse to map alpha-synuclein topography throughout the adult mouse brain, spinal cord, retina, and gut. ✨Pretty images below: nature.com/articles/s4153…
🧠🚨Our lab is hiring! Please spread the word. Multiple exciting projects in the lab including a fully funded postdoctoral fellowship/research associate position. @uOttawa @uOttawaBMRI @ALSCanada @CAN_ACN

United States Trends
- 1. Doran 37.4K posts
- 2. #Worlds2025 72K posts
- 3. #T1WIN 38.5K posts
- 4. Faker 52.4K posts
- 5. Good Sunday 53.9K posts
- 6. Silver Scrapes 4,164 posts
- 7. Guma 9,622 posts
- 8. #sundayvibes 3,871 posts
- 9. #T1fighting 4,063 posts
- 10. Keria 16.4K posts
- 11. O God 7,677 posts
- 12. Blockchain 198K posts
- 13. Oner 14K posts
- 14. #SundayMorning 1,536 posts
- 15. Faye 51.9K posts
- 16. Option 2 4,558 posts
- 17. Vergil 9,073 posts
- 18. OutKast 24.2K posts
- 19. Boots 30.1K posts
- 20. Lubin 6,046 posts
You might like
-
 uOttawaCMM
uOttawaCMM
@OttawaCmm -
 Greg Silasi
Greg Silasi
@gsilasi -
 uOttawaBMI
uOttawaBMI
@uOttawaBMI -
 S.ChenLab uOttawa
S.ChenLab uOttawa
@SChenlab -
 Paul Marcogliese
Paul Marcogliese
@PCMarcogliese -
 Tabrez J. Siddiqui
Tabrez J. Siddiqui
@SiddiquiLab -
 Tuan Bui
Tuan Bui
@TuanBuiLab -
 Dr. Anastassia Voronova
Dr. Anastassia Voronova
@AMVoronova -
 Karun K Singh
Karun K Singh
@karunsinghneuro -
 Daniel Figeys
Daniel Figeys
@DanielFigeys -
 Mireille Khacho
Mireille Khacho
@MireilleKhacho -
 Khaled Abdelrahman
Khaled Abdelrahman
@K_S_Abdelrahman -
 Konrad Ricke
Konrad Ricke
@KonradRicke
Something went wrong.
Something went wrong.