SPAdes assembler
@spadesassembler
You might like
Meet hpcSPAdes - the same assembler reworked for computing clusters! Try it out in our latest 4.2 release: github.com/ablab/spades
Despite our intentions to halt active SPAdes development, the new SPAdes 4.1 is here github.com/ablab/spades Improved performance and some important fixes.
This preprint is a draft. But given the immense utility of Logan for biology and medicine, we are immediately/freely releasing the data for early-adoption. Reach out if you’d like to collaborate. #Genomics is our shared evolutionary heritage, and so we must keep it free.

While analyzing millions of gigabytes of data may seem insurmountable today, at one point so was analyzing an entire human genome. Open source #bioinformatics illuminated the #genomic era, and now we invite the community to enter the #petabase era together.
To accomplish petabase-scale sequence assembly, we deployed a massive #AWS Batch #cloudcomputing workflow, reaching peak simultaneous-usage of 2.18 million vCPUs. After a year of preparation, the runs were executed in only 30 hours.

Logan is the assembly of all 27M DNA/RNA-seq data (44+ petabases). It expands the available genetic diversity across the Tree of Life by orders of magnitude. And it enabled us to perform a true pan-SRA sequence search in only 11 hours. Data link (cc0): registry.opendata.aws/pasteur-logan/

Today we’re excited to freely share an early-version of, perhaps, the world’s most expansive genetics dataset: Logan. #bioinformatics #petabase #genetics #genomics #openscience biorxiv.org/content/10.110…
SPAdes 4.0 is released! github.com/ablab/spades/r… Since 2022 SPAdes is not related to any institution and is maintained by the SPAdes team - an international group of volunteers. Although this might be the last major release, we strongly intend to continue supporting our users!
SPAdes 4.0 is released! github.com/ablab/spades/r… Since 2022 SPAdes is not related to any institution and is maintained by the SPAdes team - an international group of volunteers. Although this might be the last major release, we strongly intend to continue supporting our users!
SPAdes 3.15.5 is released. Support for DNA HMMs and couple bug-fixes. Update @ github.com/ablab/spades/r…
BinSPreader: refine binning results for fuller MAG reconstruction biorxiv.org/cgi/content/sh… #biorxiv_bioinfo
SPAdes 3.15.4 is released! A bunch of bug-fixes, including fixes for new MacOS and Python 3.10. Consider updating @ cab.spbu.ru/software/spade… or github.com/ablab/spades/r…
Serratus is published in @Nature!🎉 Planetary viral discovery by analysis of all available RNA sequencing data. nature.com/articles/s4158…

And @RayanChikhi wrote the short summary a while ago:
Serratus is now published in @Nature :) nature.com/articles/s4158… We searched 5.7M seq libraries (10.2 petabases) for all 15,000 known RNA viruses. In 11 days, we uncovered 130,000+ new RNA viruses (incl 9 new CoV, with a twist). That’s near an order of magnitude bump. [1/N] 🧵👇
![hackseq's tweet image. Serratus is now published in @Nature :) nature.com/articles/s4158…
We searched 5.7M seq libraries (10.2 petabases) for all 15,000 known RNA viruses. In 11 days, we uncovered 130,000+ new RNA viruses (incl 9 new CoV, with a twist). That’s near an order of magnitude bump.
[1/N] 🧵👇](https://pbs.twimg.com/media/FKCqFhNXwAQ-DfJ.jpg)
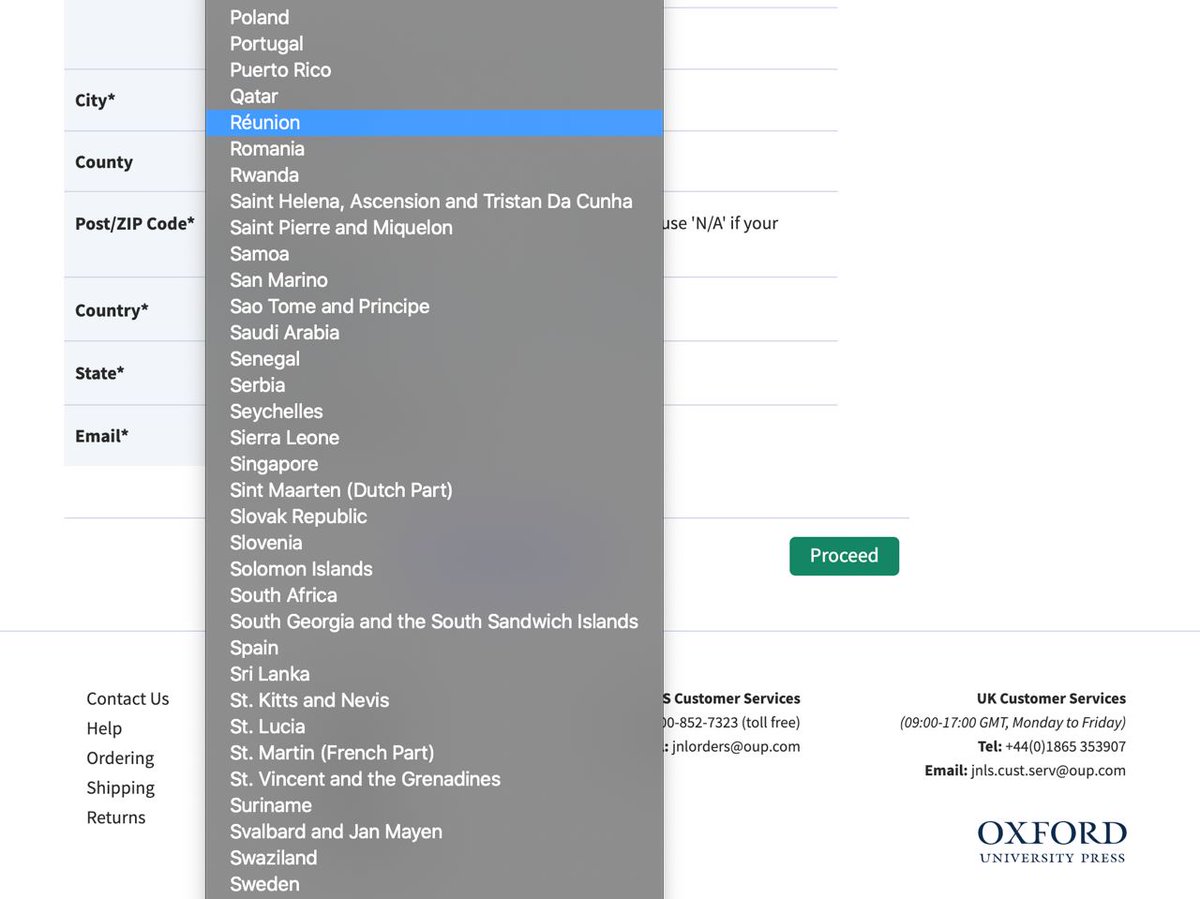
Apparently Russia does not exist for @OxUniPress. As you can see, there is no way to select Russia in Correspondence / Billing form, so one cannot pay page charges and proceed with coronaSPAdes publication. @OxfordJournals will you please comment on reason of such discrimination?
We are pleased to announce that @spadesassembler and @quast_bioinf have received grant from @cziscience to further support, enhance and develop SPAdes and QUAST!
🚨#OpenSource tools are the invisible foundation of biomedicine. That’s why we’re continuing to support open source software projects essential for scientists. Learn more about our grantees czi.co/3kH2Hpr
The last invited talk at #BiATA2021 today is about Sequence-based Approaches to Natural Product Discovery by Paul Jensen youtube.com/watch?v=w9pcHf…

youtube.com
YouTube
Sequence-based Approaches to Natural Product Discovery | Paul Jensen
Time for Computational Methods for Comparison and Consensus of Tumor Evolution Trees by @lkoesper #BiATA2021 youtube.com/watch?v=-0NZ5Y…

youtube.com
YouTube
Computational Methods for Comparison and Consensus of Tumor Evoluti...
And now at #BiATA2021 Sara Saheb Kashaf is talking about expanding our understanding of the human skin microbiome diversity and functions using genome-resolved metagenomics. youtube.com/watch?v=_zTi7B…

youtube.com
YouTube
Expanding our understanding of the human skin microbiome diversity |...
#BiATA2021 starts with the invited talk of Nikolai Ravin about metagenomic studies of deep subsurface environments. youtu.be/X_EqdIniOtk
youtube.com
YouTube
Metagenomic studies of deep subsurface environments | Nikolai Ravin
United States Trends
- 1. #River 5,813 posts
- 2. Jokic 28.1K posts
- 3. Namjoon 69.5K posts
- 4. Lakers 52.3K posts
- 5. FELIX VOGUE COVER STAR 9,270 posts
- 6. #FELIXxVOGUEKOREA 9,607 posts
- 7. #FELIXxLouisVuitton 8,909 posts
- 8. Rejoice in the Lord 1,209 posts
- 9. #ReasonableDoubtHulu N/A
- 10. #AEWDynamite 51.7K posts
- 11. Simon Nemec 2,378 posts
- 12. Clippers 15.2K posts
- 13. Shai 16.4K posts
- 14. Mikey 66.4K posts
- 15. Thunder 39.6K posts
- 16. Rory 8,378 posts
- 17. Ty Lue 1,290 posts
- 18. New Zealand 14.6K posts
- 19. Visi 7,798 posts
- 20. Valve 63K posts
You might like
Something went wrong.
Something went wrong.