Yuchun Guo
@yguo2k
Computational Biology Scientist in Biotech, Greater Boston. interested in #MachineLearning, #GeneRegulation, #RNA, anti-sense oligo
Bạn có thể thích
Excited to share our new paper “JMnorm: a novel joint multi-feature normalization method for integrative and comparative epigenomics”! Building cross-cell-type predictive models with epigenomic features? This is the method for you. Code in GitHub. academic.oup.com/nar/article/do…
Today we’re releasing an alpha version of RibonanzaNet2. Rnet2 is a 100M-parameter foundation model for #RNA structure, trained on chemical mapping profiles for 30M RNAs with complex structure. kaggle.com/c/stanford-rna… (1/N)
Are RNA 3D models accurate enough to be biologically useful? #CASP16 experimentalists say: not so much. doi.org/10.1101/2025.0… Thanks for leading this paper @rachaelkretsch @AKryshtafovych! 1/2
Our CFO Kelly Gold and Co-Founders Richard Young & Leonard Zon spoke with @ZachinBoston about our work in #regRNA biology to upregulate #GeneExpression and potentially correct protein deficiencies underlying many genetic diseases. More in this @MIT article:bit.ly/4iBnzL2
Announcing the new Pseudogenes track for hg38! The composite track contains pseudogene predictions and their corresponding parent genes, as identified by PseudoPipe. This release includes the Pseudogene Parents and Pseudogenes tracks. Learn more at genome.ucsc.edu/goldenPath/new….

I'm honored to be inducted as a fellow into the American Institute for Medical and Biological Engineering @aimbe. Had a great visit to Capitol Hill and advocated for biomedical engineering research. @NU_BMG_SQE @NUFeinbergMed


#RNA structure prediction remains unsolved -- nice tech feature incl #deeplearning and #CASP16 in @Nature nature.com/articles/d4158…
Applying for a Bioinformatics position? Your CV competes with hundreds of others. Here’s how to stand out, based on my hiring experience: 🧵👇
Diverse Database and Machine Learning Model to narrow the generalization gap in RNA structure prediction bioRxiv biorxiv.org/cgi/content/sh…
Existing methods normalize epigenetic features independently and distort relationships among the features. JMnorm jointly normalizes multiple features, enables data sharing and preserves the relationships. It works even with only two features, e.g. ATAC and H3K27ac.
Excited to share our new paper “JMnorm: a novel joint multi-feature normalization method for integrative and comparative epigenomics”! Building cross-cell-type predictive models with epigenomic features? This is the method for you. Code in GitHub. academic.oup.com/nar/article/do…
Nature research paper: Cell type directed design of synthetic enhancers go.nature.com/3RkatGn
#ICYMI: CAMP4’s scientific founders recently shared their insights on #regRNAs, the foundation of our novel science. 🏔️Watch the highlights to learn about the potential of our game-changing approach in #genetic diseases: ow.ly/Vm2c50PIKKu
We are thrilled to share our manuscript ‘An RNA foundation model enables discovery of disease mechanisms and candidate therapeutics’ as a preprint! 🧵 biorxiv.org/content/10.110…
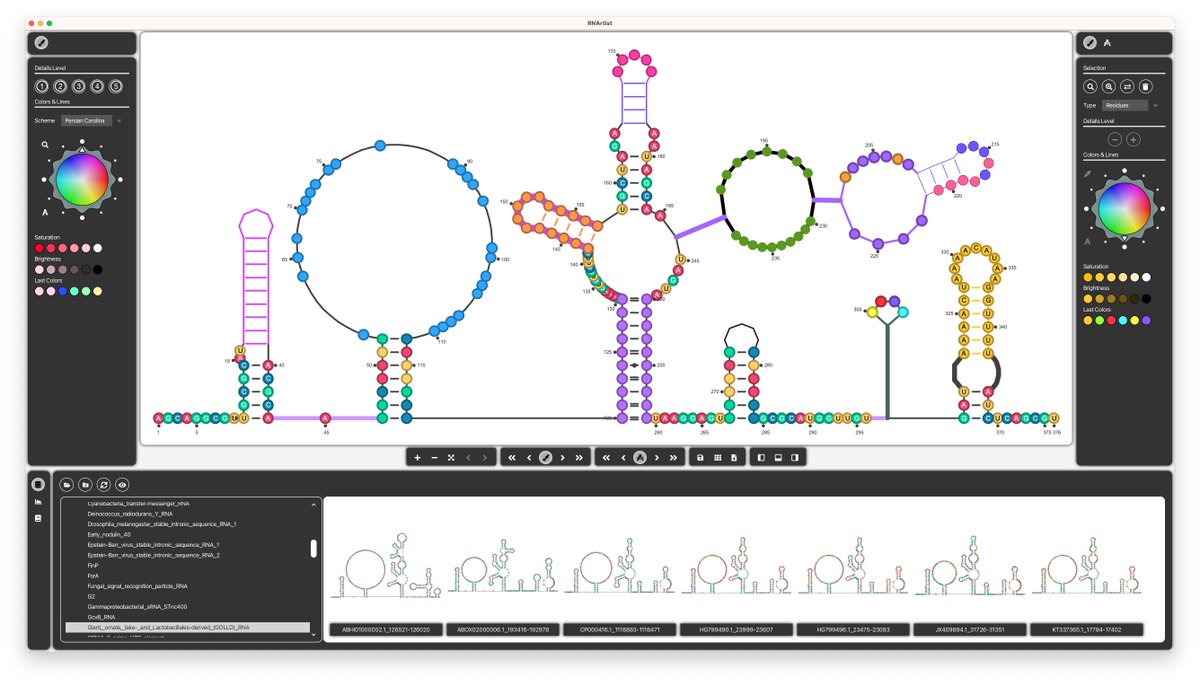
You can now give a try to RNArtist for MacOS/Linux (Windows next week). This project is under active development: it is not perfect and many exciting features will come soon. All details here: github.com/fjossinet/RNAr… #bioinformatics #RNA #visualization #java #javafx #kotlin
We’ve teamed up with @kaggle and the global #deeplearning community to accelerate progress in #RNA structure prediction. Check out the new #Ribonanza challenge: kaggle.com/c/stanford-rib… 1/7
😲 Time to revise textbooks 🤔 Title: "Transcription factors interact with RNA to regulate genes" "at least half of TFs also bind RNA, doing so through a previously unrecognized domain" cell.com/molecular-cell…
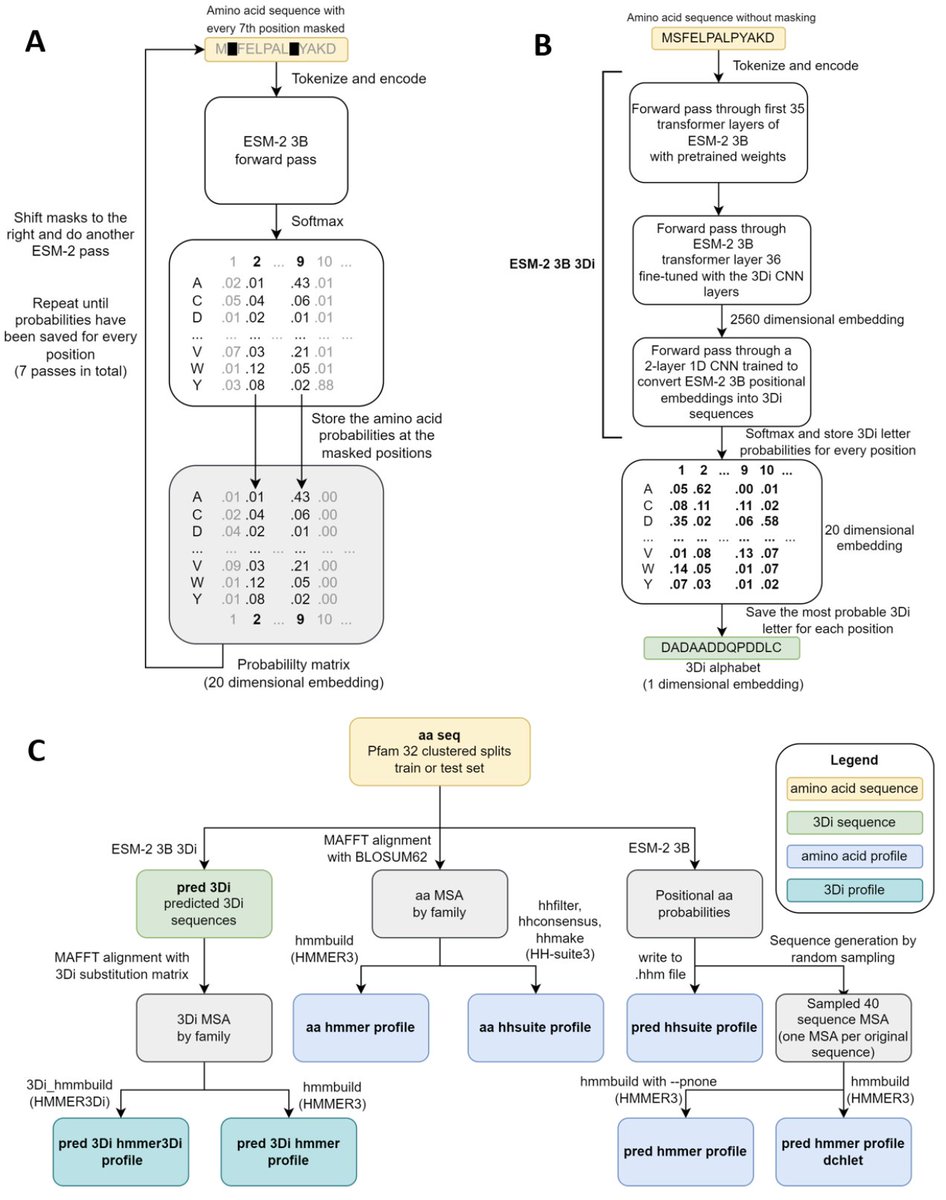
"Sensitive remote homology search by local alignment of small positional embeddings from protein language models" ESM2 embeddings used " as input to the highly optimized Foldseek, HMMER3, and HH-suite search algorithms" biorxiv.org/content/10.110…
Delighted to share a short review article published in @ScienceAdvances!! Enhancer dynamics: Unraveling the mechanism of transcriptional bursting. science.org/doi/10.1126/sc…

Read this from @WhiteheadInst on findings from CAMP4 co-founder @youngricka’s lab, published in @MolecularCell, showing that RNA binds to transcription factors to regulate #geneexpression, providing biological validation to CAMP4’s approach. #RNADay
“We show that RNA binding by transcription factors is a general phenomenon,” postdocs Ozgur Oksuz says. New research shows that many transcription factors also bind to RNA—an insight that may lead to a better understanding of gene regulation. wi.mit.edu/news/it-takes-…

All talks from the main #RECOMB2023 meeting are now available at: youtube.com/playlist?list=… Also: Day 1: youtube.com/playlist?list=… Day 2: youtube.com/playlist?list=… Day 3: youtube.com/playlist?list=… Day 4: youtube.com/playlist?list=…
United States Xu hướng
- 1. Branch 37.3K posts
- 2. Chiefs 112K posts
- 3. Red Cross 54.1K posts
- 4. Lions 89.7K posts
- 5. #njkopw 8,408 posts
- 6. Exceeded 5,856 posts
- 7. Binance DEX 5,145 posts
- 8. Mahomes 34.9K posts
- 9. Rod Wave 1,674 posts
- 10. Air Force One 58.1K posts
- 11. #LaGranjaVIP 83.5K posts
- 12. Eitan Mor 17.6K posts
- 13. #TNABoundForGlory 59.9K posts
- 14. #LoveCabin 1,389 posts
- 15. Ziv Berman 20.7K posts
- 16. Alon Ohel 18.2K posts
- 17. Tel Aviv 59.8K posts
- 18. Matan Angrest 16.4K posts
- 19. Omri Miran 16.4K posts
- 20. Bryce Miller 4,612 posts
Bạn có thể thích
-
 Julia Zeitlinger (@[email protected])
Julia Zeitlinger (@[email protected])
@JuliaZeitlinger -
 Ye Zheng
Ye Zheng
@yezhengSTAT -
 Nezar Abdennur
Nezar Abdennur
@nv1ctus -
 Raluca Gordan
Raluca Gordan
@RalucaGordan -
 Luca Pinello
Luca Pinello
@lucapinello -
 Philipp Maass
Philipp Maass
@NucleomeHunter -
 Sourya Bhattacharyya
Sourya Bhattacharyya
@SouryaBhattach5 -
 Ross Hardison
Ross Hardison
@rosshardison -
 Gürkan Yardımcı
Gürkan Yardımcı
@Gurkan_Yardimci -
 Eric Guo
Eric Guo
@ericyguo
Something went wrong.
Something went wrong.