#diffdock search results
Discover how DiffDock can simplify molecular docking. Just upload your molecule and target protein, adjust a few settings, and get accurate results. A powerful tool for researchers, made easy to use. #DiffDock #NVIDIANIM #NVIDIAhealthcare Try today ➡️ nvda.ws/4gffH0G
Check out @nvidia's BioNeMo api that makes it easy to do virtual screening with #DiffDock and #ESMFold

MIT researchers built DiffDock, a model that may one day be able to find new drugs faster than traditional methods and reduce the potential for adverse side effects. #diffdock #research #diffusionmodel #science #newdrug #techexplorist

want to play with #DiffDock? check out this 🤗/Gradio web app demoing DiffDock from the @BarzilayRegina and Jaakola labs in @AIHealthMIT huggingface.co/spaces/reginab…

Use DiffDock as web application #DiffDock #cheminformatics #memo iwatobipen.wordpress.com/2024/08/24/use…
MIT researchers develop #DiffDock drug discovery model to quickly identify potential drug molecules w/ fewer adverse side effects. Could speed drug dev & reduce clinical trial failure rate #DrugDiscovery #AlphaFold2 💊🤞 #MITNews
AI models that predict protein-protein and protein-ligand interactions have finally met their match. #PINDER #PLINDER #DiffDock bit.ly/4ccU2nG
Semester may be over, but our research on drug design @MissouriState is still going strong! My student Gaige Riggs & I appreciate the discussion w/ @HannesStaerk on #Diffdock, the new #AI-based molecular docking code development, led by @GabriCorso fr. Prof. Jaakkola's group @MIT

Optimizing drug discovery with generative diffusion models @MIT #Protein #drug #DIFFDock #research techexplorist.com/optimizing-dru…
Use DiffDock as web application #DiffDock #cheminformatics #memo iwatobipen.wordpress.com/2024/08/24/use… @iwatobipenから
MIT researchers built DiffDock machine-learning model to find new drugs faster #MIT #DiffDock #machinelearning #drugdiscovery #pharma #ai #generativemodels #healthtech #pharmatech #medtech #medtechnews #science moremedtech.com/mit-researcher…

A new Machine Learning Engineer position is open in our group! Come to work with us on developing open-source AI tools for drug discovery and biomedicine such as #DiffDock, #FrameDiff, #RFDiffusion, and many more! (Please RT!)
Join us at #JameelClinic, a global hub leading in knowledge, innovation, and AI for health + biotech! If you're a machine learning engineer, we invite you to become an integral member of this dynamic ecosystem. 🌟Apply now: careers.peopleclick.com/careerscp/clie…

if you missed @GabriCorso, @HannesStaerk, and Bowen Jing's fantastic seminar about #DiffDock that happened earlier this year, you can now check out the recording 👇 youtu.be/_KBqVh6YbgI
youtube.com
YouTube
DiffDock: Diffusion Steps, Twists and Turns for Molecular Docking and...
Kudos to @GabrieleCorso, @HannesStark for their excellent work on Docking ("DiffDock"). Here is the link for revised Colab Notebook - which allows you to run docking for a given input protein and ligand. Hope you find it useful. github.com/suneelbvs/Diff… #compchem #DiffDock
Congratulations to @GabriCorso, @HannesStaerk, Bowen Jing, @BarzilayRegina and Tommi Jaakkola - and everyone at the MIT Jameel Clinic 🙏 #DiffDock
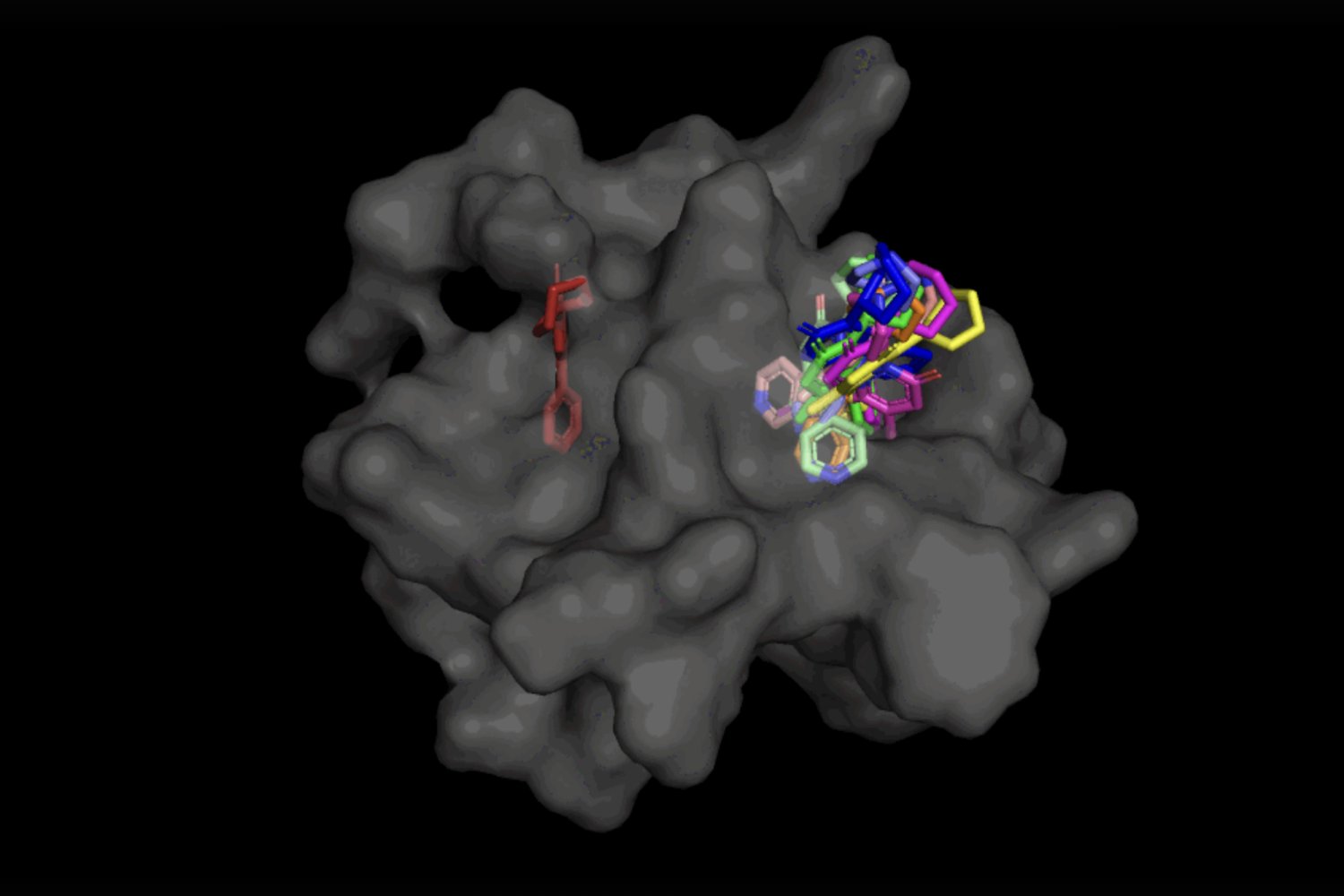
With its ability to generate many different potential protein-to-ligand (drug molecule) poses at once, DiffDock is about to change the game for drug discovery thanks to #JameelClinic researchers: news.mit.edu/2023/speeding-…
CDD Vault Update (April 2025 #3): AI+ Folding & Docking More details in our latest blog post:collaborativedrug.pulse.ly/cqzsbdgkw1 #CDDVault #AlphaFold2 #DiffDock #StructuralBiology #DrugDiscovery
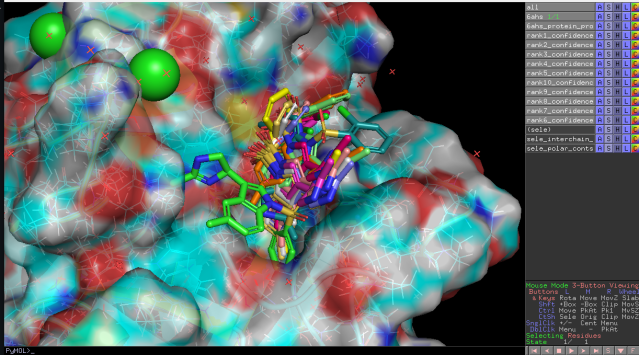
Instead of spending extra time learning various services and tools, you can find, visualize, and dock molecules on a single platform by entering text prompts in just a few minutes. See the detailed script at the link below: bit.ly/4gnPcGs #pymol #diffdock
TLDR; #DiffDock is an AI model by MIT researchers that accelerates drug discovery, predicts protein interactions, and enhances pharmaceutical research accuracy. news.mit.edu/2023/speeding-…
(4/9) Enter DiffDock! It uses diffusion generative models to generate new 3D coordinates for ligand-protein binding, and even works on proteins it's never seen before. DiffDock's "blind docking" is the future of drug design. #DiffDock #GenerativeModels
CDD Vault Update (April 2025 #3): AI+ Folding & Docking More details in our latest blog post:collaborativedrug.pulse.ly/cqzsbdgkw1 #CDDVault #AlphaFold2 #DiffDock #StructuralBiology #DrugDiscovery
Instead of spending extra time learning various services and tools, you can find, visualize, and dock molecules on a single platform by entering text prompts in just a few minutes. See the detailed script at the link below: bit.ly/4gnPcGs #pymol #diffdock
Discover how DiffDock can simplify molecular docking. Just upload your molecule and target protein, adjust a few settings, and get accurate results. A powerful tool for researchers, made easy to use. #DiffDock #NVIDIANIM #NVIDIAhealthcare Try today ➡️ nvda.ws/4gffH0G
Use DiffDock as web application #DiffDock #cheminformatics #memo iwatobipen.wordpress.com/2024/08/24/use… @iwatobipenから
Use DiffDock as web application #DiffDock #cheminformatics #memo iwatobipen.wordpress.com/2024/08/24/use… @iwatobipenから
Use DiffDock as web application #DiffDock #cheminformatics #memo iwatobipen.wordpress.com/2024/08/24/use…
AI models that predict protein-protein and protein-ligand interactions have finally met their match. #PINDER #PLINDER #DiffDock bit.ly/4ccU2nG
want to play with #DiffDock? check out this 🤗/Gradio web app demoing DiffDock from the @BarzilayRegina and Jaakola labs in @AIHealthMIT huggingface.co/spaces/reginab…

Check out @nvidia's BioNeMo api that makes it easy to do virtual screening with #DiffDock and #ESMFold

Semester may be over, but our research on drug design @MissouriState is still going strong! My student Gaige Riggs & I appreciate the discussion w/ @HannesStaerk on #Diffdock, the new #AI-based molecular docking code development, led by @GabriCorso fr. Prof. Jaakkola's group @MIT

if you missed @GabriCorso, @HannesStaerk, and Bowen Jing's fantastic seminar about #DiffDock that happened earlier this year, you can now check out the recording 👇 youtu.be/_KBqVh6YbgI

youtube.com
YouTube
DiffDock: Diffusion Steps, Twists and Turns for Molecular Docking and...
A new Machine Learning Engineer position is open in our group! Come to work with us on developing open-source AI tools for drug discovery and biomedicine such as #DiffDock, #FrameDiff, #RFDiffusion, and many more! (Please RT!)
Join us at #JameelClinic, a global hub leading in knowledge, innovation, and AI for health + biotech! If you're a machine learning engineer, we invite you to become an integral member of this dynamic ecosystem. 🌟Apply now: careers.peopleclick.com/careerscp/clie…

TLDR; #DiffDock is an AI model by MIT researchers that accelerates drug discovery, predicts protein interactions, and enhances pharmaceutical research accuracy. news.mit.edu/2023/speeding-…
(4/9) Enter DiffDock! It uses diffusion generative models to generate new 3D coordinates for ligand-protein binding, and even works on proteins it's never seen before. DiffDock's "blind docking" is the future of drug design. #DiffDock #GenerativeModels
MIT researchers built DiffDock, a model that may one day be able to find new drugs faster than traditional methods and reduce the potential for adverse side effects. #diffdock #research #diffusionmodel #science #newdrug #techexplorist

Optimizing drug discovery with generative diffusion models @MIT #Protein #drug #DIFFDock #research techexplorist.com/optimizing-dru…
Congratulations to @GabriCorso, @HannesStaerk, Bowen Jing, @BarzilayRegina and Tommi Jaakkola - and everyone at the MIT Jameel Clinic 🙏 #DiffDock
With its ability to generate many different potential protein-to-ligand (drug molecule) poses at once, DiffDock is about to change the game for drug discovery thanks to #JameelClinic researchers: news.mit.edu/2023/speeding-…
Kudos to @GabrieleCorso, @HannesStark for their excellent work on Docking ("DiffDock"). Here is the link for revised Colab Notebook - which allows you to run docking for a given input protein and ligand. Hope you find it useful. github.com/suneelbvs/Diff… #compchem #DiffDock
Check out @nvidia's BioNeMo api that makes it easy to do virtual screening with #DiffDock and #ESMFold

want to play with #DiffDock? check out this 🤗/Gradio web app demoing DiffDock from the @BarzilayRegina and Jaakola labs in @AIHealthMIT huggingface.co/spaces/reginab…

MIT researchers built DiffDock, a model that may one day be able to find new drugs faster than traditional methods and reduce the potential for adverse side effects. #diffdock #research #diffusionmodel #science #newdrug #techexplorist

Semester may be over, but our research on drug design @MissouriState is still going strong! My student Gaige Riggs & I appreciate the discussion w/ @HannesStaerk on #Diffdock, the new #AI-based molecular docking code development, led by @GabriCorso fr. Prof. Jaakkola's group @MIT

MIT researchers built DiffDock machine-learning model to find new drugs faster #MIT #DiffDock #machinelearning #drugdiscovery #pharma #ai #generativemodels #healthtech #pharmatech #medtech #medtechnews #science moremedtech.com/mit-researcher…

Something went wrong.
Something went wrong.
United States Trends
- 1. Comey 54.6K posts
- 2. Thanksgiving 156K posts
- 3. Jimmy Cliff 28.5K posts
- 4. #WooSoxWishList 5,230 posts
- 5. Lindsey Halligan 10.3K posts
- 6. #IDontWantToOverreactBUT 1,453 posts
- 7. #NutramentHolidayPromotion N/A
- 8. DISMISSED 27.7K posts
- 9. Pentagon 10.6K posts
- 10. The Department of War 12.7K posts
- 11. #MondayMotivation 14.5K posts
- 12. Sen. Mark Kelly 17.3K posts
- 13. DOGE 243K posts
- 14. #stayselcaday 4,811 posts
- 15. TOP CALL 5,255 posts
- 16. Hal Steinbrenner N/A
- 17. Monad 183K posts
- 18. Victory Monday 5,332 posts
- 19. AI Alert 3,435 posts
- 20. Justin Tucker N/A