#expressionandquantification نتائج البحث
MINTmap – profiling nuclear and mitochondrial… bit.ly/2lJ99iZ #ExpressionandQuantification #OtherTools #ReaderConributions #RNAseq
How to count multi-mapping reads?: RNA-Seq is… bit.ly/2xaqGXG #ExpressionandQuantification #featureCounts #geneexpression #RNAseq
Gene-level false discovery rate control for… bit.ly/2gTrmd9 #ExpressionandQuantification #UserSubmittedPosts #RNAseq
voomDDA: discovery of diagnostic biomarkers and… bit.ly/2gnPilP #ExpressionandQuantification #UserSubmittedPosts #RNAseq
powsimR – power analysis for bulk and single cell… bit.ly/2gPLb2u #ExpressionandQuantification #StatisitcalAnalysis #RNAseq
Single molecule counting and assessment of random… bit.ly/2fJkDz5 #ExpressionandQuantification #molecularbarcoding #RNASeq #RNAseq
Network embedding-based representation learning for… bit.ly/2xVShfC #ExpressionandQuantification #dimensionalreduction #RNAseq
Improved transcript discovery from partially observed… bit.ly/2w2xAud #ExpressionandQuantification #OtherTools #isoform #RNAseq
SparseDC – a sparse differential clustering algorithm for… bit.ly/2hO1ArW #DataVisualization #ExpressionandQuantification #RNAseq
Bayesian estimation of differential transcript usage from… bit.ly/2z9uIk1 #ExpressionandQuantification #alternativesplicing #RNAseq
REQUIEM – RElative QUantitation Inferred by Evaluating Mixtures:… bit.ly/2z2XyT7 #ExpressionandQuantification #accuracy #RNAseq
DEWE – Differential Expression Workflow Executor for #RNA-Seq: DEWE (sing-group.org/dewe/) is an open source application for easily executing Differential Expression analyses in #RNA-Seq data. DEWE… bit.ly/2n63PFC #ExpressionandQuantification #Uncategorized #RNAseq
Assessment of data transformations for model-based clustering of #RNA-Seq data: Quality control, global biases, normalization, and analysis methods for #RNA-Seq data are quite… bit.ly/2CStYxu #ExpressionandQuantification #StatisitcalAnalysis #Clustering #RNAseq
Strawberry – Fast and accurate genome-guided transcript reconstruction and quantification from #RNA-Seq: Iowa State University researchers propose a novel method and software tool,… bit.ly/2ioUDh6 #ExpressionandQuantification #algorithm #geneexpression #RNAseq
APAtrap – identification and quantification of alternative polyadenylation sites from #RNA-seq data: Alternative polyadenylation (APA) has been increasingly recognized as a crucial mechanism that… bit.ly/2mSE7nA #ExpressionandQuantification #OtherTools #RNAseq
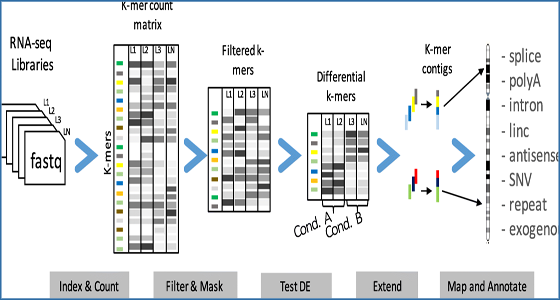
DE-kupl: exhaustive capture of biological variation in #RNA-seq data through k-mer decomposition: University of Montpellier researchers have developed a k-mer-based computational protocol, DE-kupl,… bit.ly/2DR49yO #ExpressionandQuantification #OtherTools #DEkupl #RNAseq
Hierarchical analysis of #RNA-Seq reads improves the accuracy of allele-specific expression: Allele-specific expression (ASE) refers to the differential abundance of the allelic copies… bit.ly/2o5FgcY #ExpressionandQuantification #allelespecificexpression #EMASE #RNAseq
RNentropy – an entropy-based tool for the detection of significant variation of gene expression across multiple #RNA-Seq experiments: RNA sequencing (RNA-Seq) has become the experimental standard… bit.ly/2sdNy73 #ExpressionandQuantification #StatisitcalAnalysis #RNAseq
Overcome analytical bottlenecks in experiments with limited number of replicates and low sequencing coverage: In current statistical methods for calling differentially expressed genes in #RNA-Seq… bit.ly/2E5ekj5 #ExpressionandQuantification #StatisitcalAnalysis #RNAseq
Quartz-Seq2 – a high-throughput single-cell #RNA-sequencing method that effectively uses limited sequence reads: High-throughput single-cell #RNA-seq methods assign limited unique molecular… bit.ly/2DnzWqr #ExpressionandQuantification #Cellsorter #Flowcytometry #RNAseq
Single cell #RNA-seq data clustering using TF-IDF based methods: Single cell transcriptomics is critical for understanding cellular heterogeneity and identification of novel cell types.… bit.ly/2PtfVZj #ExpressionandQuantification #OtherTools #StatisitcalAnalysis
Quartz-Seq2 – a high-throughput single-cell #RNA-sequencing method that effectively uses limited sequence reads: High-throughput single-cell #RNA-seq methods assign limited unique molecular… bit.ly/2DnzWqr #ExpressionandQuantification #Cellsorter #Flowcytometry #RNAseq
Assessment of data transformations for model-based clustering of #RNA-Seq data: Quality control, global biases, normalization, and analysis methods for #RNA-Seq data are quite… bit.ly/2CStYxu #ExpressionandQuantification #StatisitcalAnalysis #Clustering #RNAseq
Hierarchical analysis of #RNA-Seq reads improves the accuracy of allele-specific expression: Allele-specific expression (ASE) refers to the differential abundance of the allelic copies… bit.ly/2o5FgcY #ExpressionandQuantification #allelespecificexpression #EMASE #RNAseq
RNentropy – an entropy-based tool for the detection of significant variation of gene expression across multiple #RNA-Seq experiments: RNA sequencing (RNA-Seq) has become the experimental standard… bit.ly/2sdNy73 #ExpressionandQuantification #StatisitcalAnalysis #RNAseq
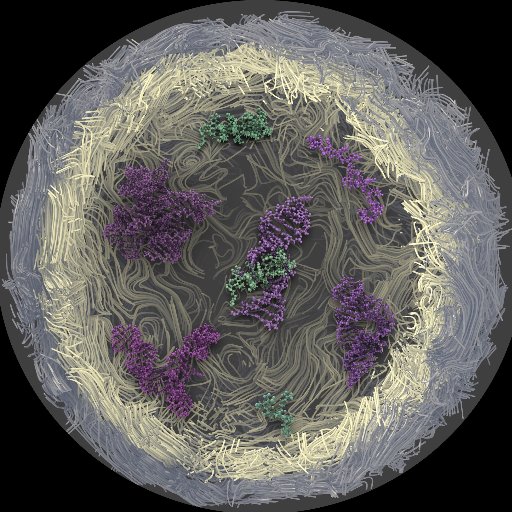
TimeLapse-seq – adding a temporal dimension to #RNA sequencing through nucleoside recoding: RNA sequencing (RNA-seq) offers a snapshot of cellular #RNA populations, but not temporal… bit.ly/2n8YBsH #ExpressionandQuantification #OtherTools #SingleMolecule #RNAseq
quanTIseq – quantifying immune contexture of human tumors: quanTIseq is the first computational pipeline for the quantification of Tumor-infiltrating Immune cells from raw #RNA-seq data and images of… bit.ly/2E40kaA #ExpressionandQuantification #haematoxylin #RNAseq
DEWE – Differential Expression Workflow Executor for #RNA-Seq: DEWE (sing-group.org/dewe/) is an open source application for easily executing Differential Expression analyses in #RNA-Seq data. DEWE… bit.ly/2n63PFC #ExpressionandQuantification #Uncategorized #RNAseq
APAtrap – identification and quantification of alternative polyadenylation sites from #RNA-seq data: Alternative polyadenylation (APA) has been increasingly recognized as a crucial mechanism that… bit.ly/2mSE7nA #ExpressionandQuantification #OtherTools #RNAseq
Overcome analytical bottlenecks in experiments with limited number of replicates and low sequencing coverage: In current statistical methods for calling differentially expressed genes in #RNA-Seq… bit.ly/2E5ekj5 #ExpressionandQuantification #StatisitcalAnalysis #RNAseq
DE-kupl: exhaustive capture of biological variation in #RNA-seq data through k-mer decomposition: University of Montpellier researchers have developed a k-mer-based computational protocol, DE-kupl,… bit.ly/2DR49yO #ExpressionandQuantification #OtherTools #DEkupl #RNAseq
SalmonTE – An ultra-fast and scalable quantification pipeline for transposable elements from next generation sequencing data: Transposable elements (TEs) are DNA sequences which are capable of… bit.ly/2kYFyUd #ExpressionandQuantification #BaylorCollegeofMedicine #RNAseq
Strawberry – Fast and accurate genome-guided transcript reconstruction and quantification from #RNA-Seq: Iowa State University researchers propose a novel method and software tool,… bit.ly/2ioUDh6 #ExpressionandQuantification #algorithm #geneexpression #RNAseq
SparseDC – a sparse differential clustering algorithm for… bit.ly/2hO1ArW #DataVisualization #ExpressionandQuantification #RNAseq
Bayesian estimation of differential transcript usage from… bit.ly/2z9uIk1 #ExpressionandQuantification #alternativesplicing #RNAseq
REQUIEM – RElative QUantitation Inferred by Evaluating Mixtures:… bit.ly/2z2XyT7 #ExpressionandQuantification #accuracy #RNAseq
powsimR – power analysis for bulk and single cell… bit.ly/2gPLb2u #ExpressionandQuantification #StatisitcalAnalysis #RNAseq
voomDDA: discovery of diagnostic biomarkers and… bit.ly/2gnPilP #ExpressionandQuantification #UserSubmittedPosts #RNAseq
Network embedding-based representation learning for… bit.ly/2xVShfC #ExpressionandQuantification #dimensionalreduction #RNAseq
Single molecule counting and assessment of random… bit.ly/2fJkDz5 #ExpressionandQuantification #molecularbarcoding #RNASeq #RNAseq
Something went wrong.
Something went wrong.
United States Trends
- 1. Eagles 81.8K posts
- 2. Eagles 81.8K posts
- 3. Jalen 21.7K posts
- 4. Caleb 40.2K posts
- 5. Ben Johnson 4,165 posts
- 6. AJ Brown 2,843 posts
- 7. Black Friday 484K posts
- 8. Swift 55.6K posts
- 9. Swift 55.6K posts
- 10. Patullo 5,143 posts
- 11. Tush Push 3,858 posts
- 12. Nebraska 13.2K posts
- 13. Nahshon Wright 1,744 posts
- 14. #CHIvsPHI 1,745 posts
- 15. Lane 49.3K posts
- 16. Kevin Byard 1,521 posts
- 17. Al Michaels N/A
- 18. Philly 16.5K posts
- 19. Sydney Brown 1,224 posts
- 20. Iowa 14.6K posts