#proteinstructureprediction 검색 결과
"The major leap in structure prediction that AlphaFold has spawned does not mean that experimental techniques have suddenly become meaningless" #AlphaFold #ProteinStructurePrediction #Crystallography doi.org/10.1107/S20532…
Meta’s generative AI ESMFold uses a large language model to predict protein structures from sequences. With lightning-fast predictions of over 700 million proteins, it's revolutionizing biotech. #AIinBiotech #ProteinStructurePrediction

Revolutionizing membrane protein design with AI-powered secondary structure prediction. New tools empower the field. #MembraneProteinDesign #ProteinStructurePrediction #AIProteinDesign Details:spj.science.org/doi/10.34133/2…
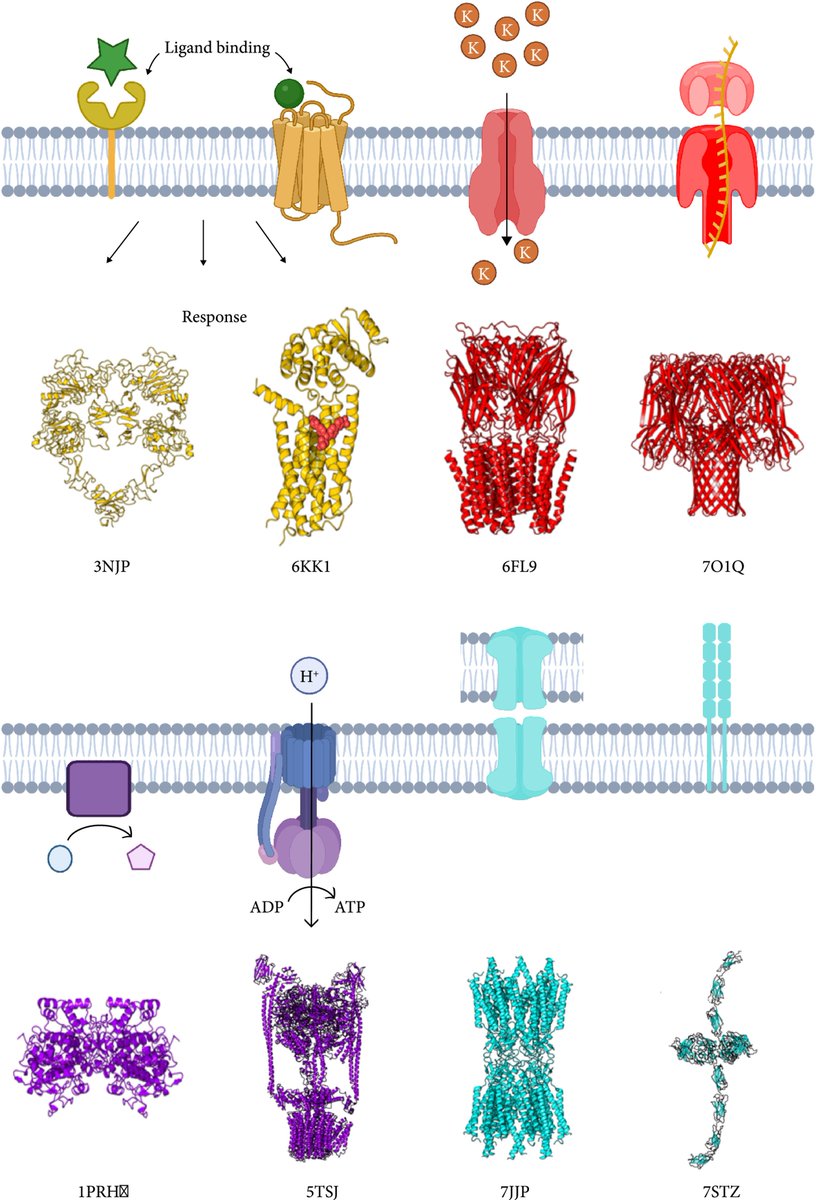
Accurate prediction of protein structures and interactions using a 3-track network biorxiv.org/content/10.110… // #DeepMind #ProteinStructurePrediction
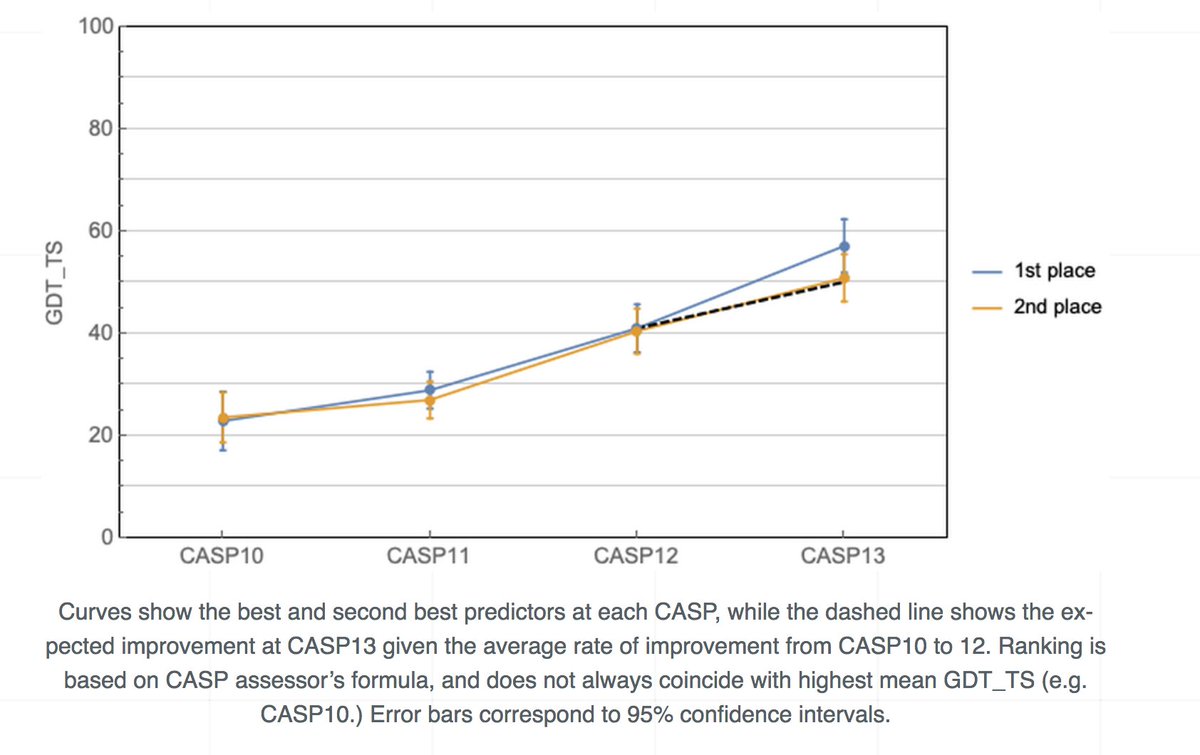
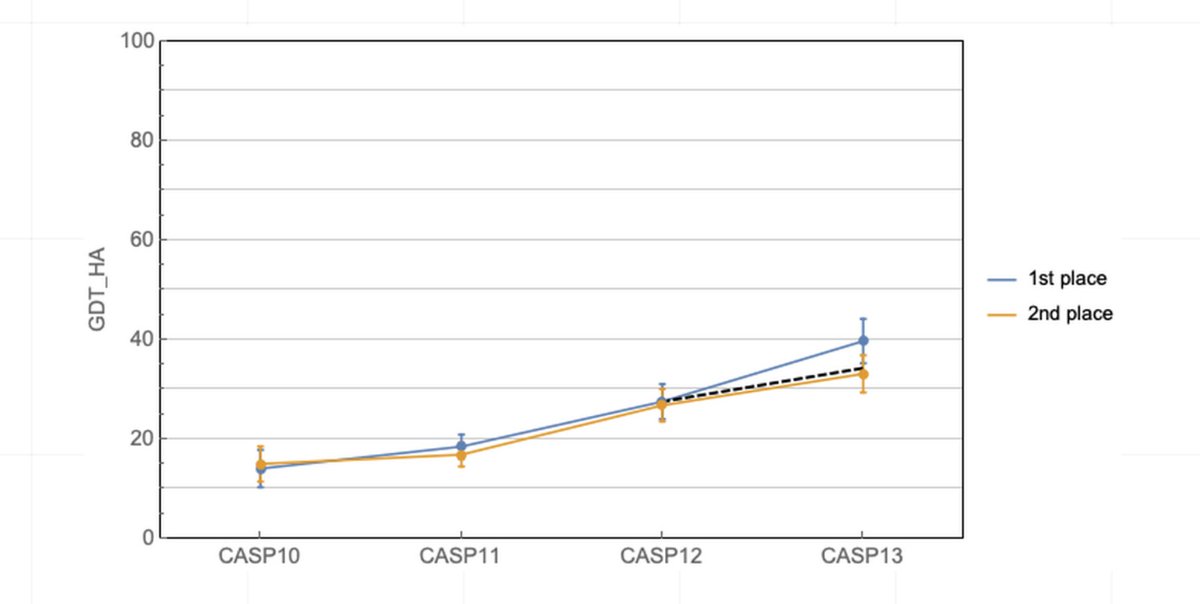
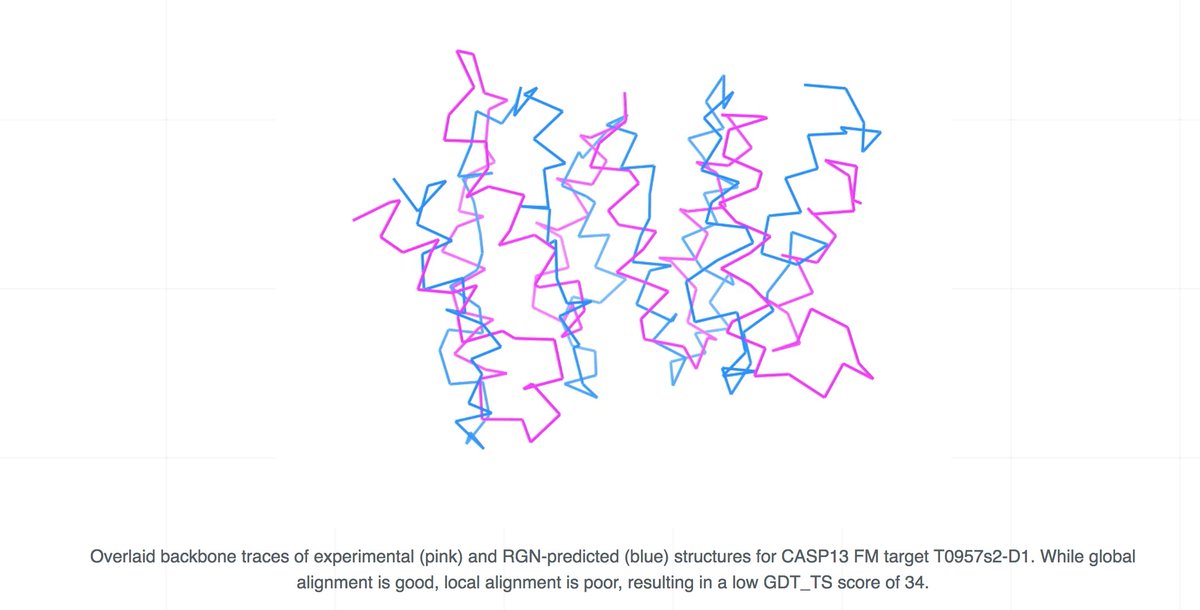
AlphaFold @ CASP13: “What just happened?” | Some Thoughts on a Mysterious Universe Blog moalquraishi.wordpress.com/2018/12/09/alp… #ProteinStructurePrediction
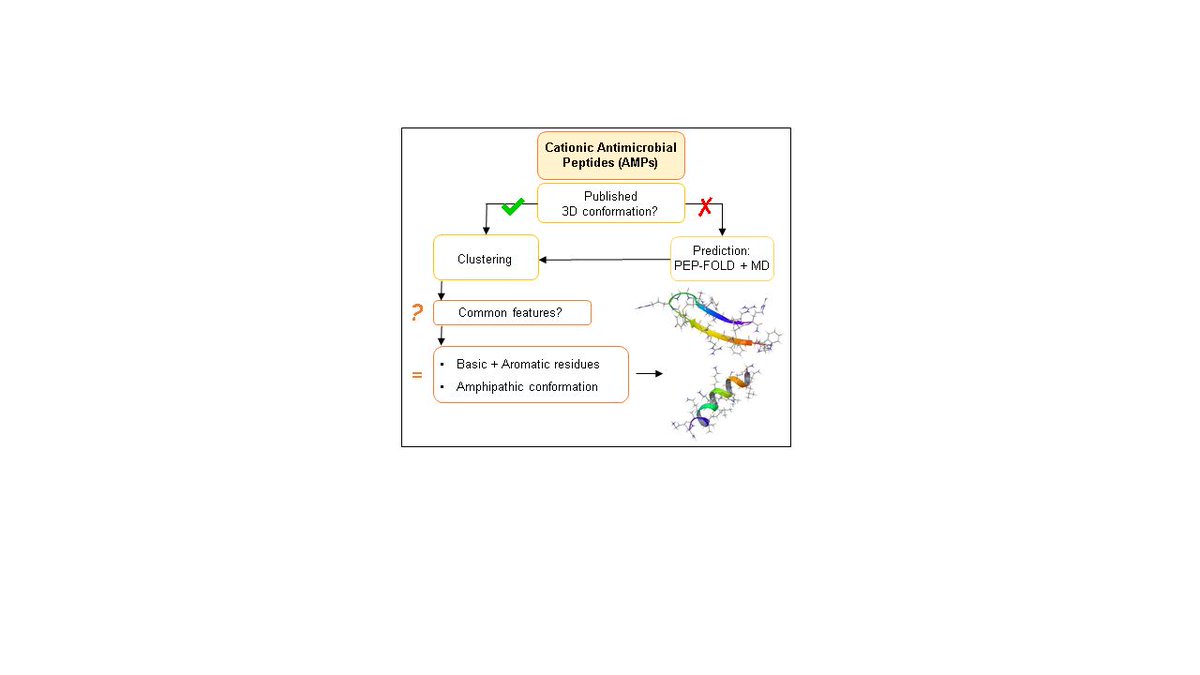
In Silico Structural Evaluation of Short Cationic Antimicrobial Peptides sci.fo/513 #MolecularDynamics #ProteinStructurePrediction @MDPIpharma
Randy J. Read et al.: AlphaFold and the future of structural biology @Cambridge_Uni @AucklandUni @uwanews @UniofOxford @ActaCrystD @IUCr #AlphaFold #ProteinStructurePrediction #Crystallography doi.org/10.1107/S20597…
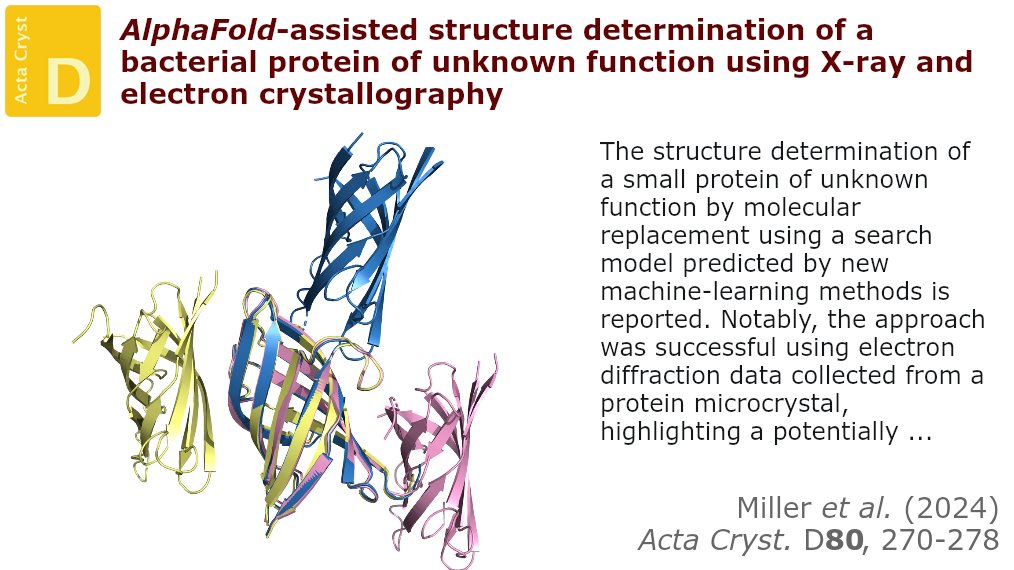
Using both X-ray and microED data, the structure of the main fragment of a protein of unkown function was solved using a predicted model of that domain as a starting point @IUCr #ElectronDiffraction #ProteinStructurePrediction #BacterialProteins doi.org/10.1107/S20597…
High-resolution de novo structure prediction from primary sequence. #ProteinStructurePrediction #Bioinformatics @biorxivpreprint biorxiv.org/content/10.110…
Just submitted last target for #CASP. Bring on the results in December! That makes a total of 426 ab initio models with our #proteinstructureprediction pipeline RBO-Aleph, #contactpredictions for 90 targets w. Epsilon-CP & 85 XL data-assisted #models robotics.tu-berlin.de/menue/research…
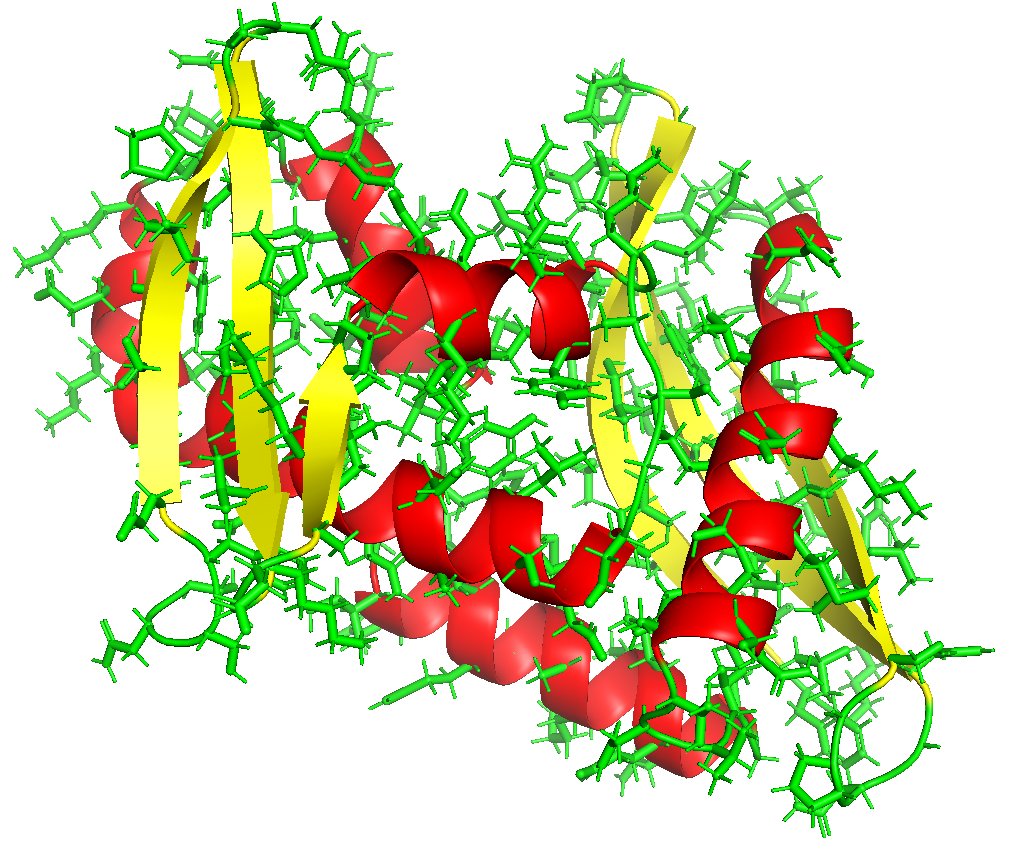
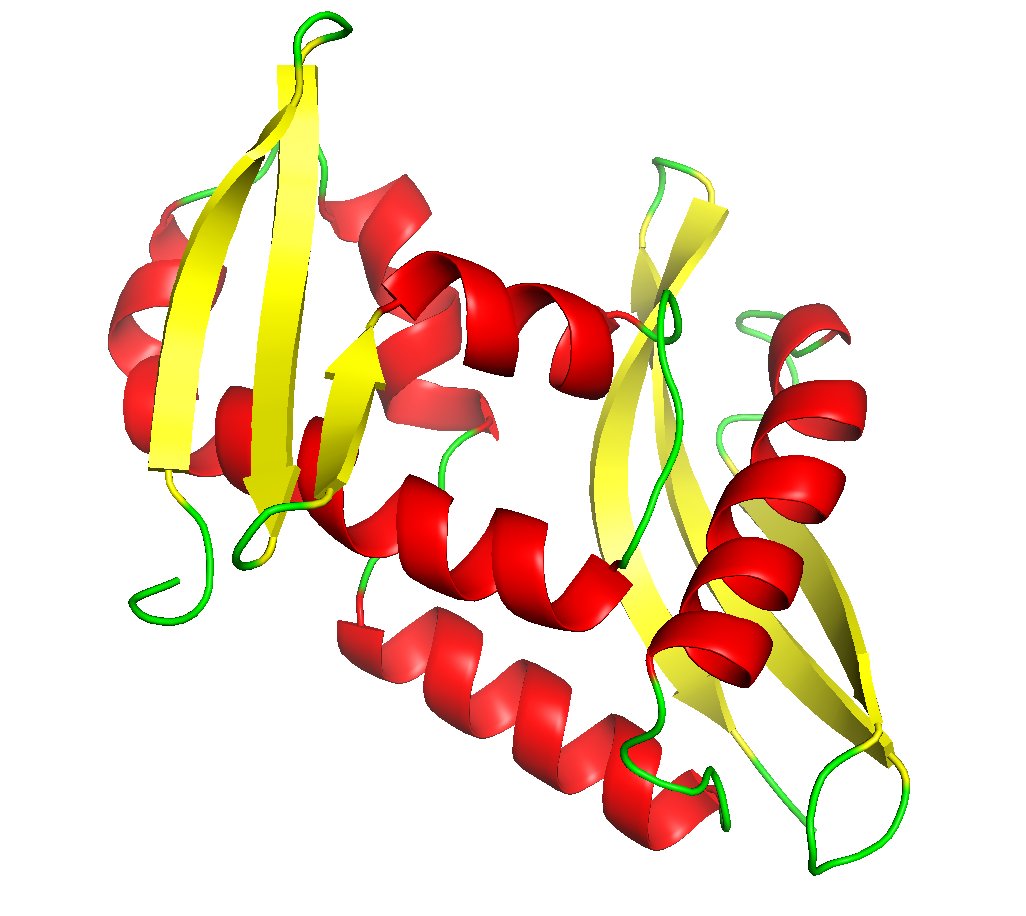
McCoy, Sammito and Read: Implications of AlphaFold2 for crystallographic phasing by molecular replacement #ProteinStructurePrediction @Cambridge_Uni ... #IUCr scripts.iucr.org/cgi-bin/paper?…
Interesting paper alert! Learn about how AlphaFold2 predicts conditionally folding regions in intrinsically disordered proteins. 🔗Check it out here: mdpi.com/2093342.. #proteinstructureprediction #AlphaFold2 #research

For #ProteinStructurePrediction lovers 👇🏻
It was great to participate today in the @NORAdotAI AlphaFold v2.0 and RoseTTAFold workshop. Experts from both teams explained their respective methods. Very enlightening 💡. Check out all recorded talks here: nora.ai/nora-webinars/…
Now both RoseTTaFold and AlphaFold2 codes are available, amazing times for #ProteinStructurePrediction 👇🏻 DeepMind’s AI for protein structure is coming to the masses nature.com/articles/d4158…
nature.com
DeepMind’s AI for protein structure is coming to the masses
Nature - Machine-learning systems from the company and from a rival academic group are now open source and freely accessible.
THIS IS a true REVOLUTION 💥 My deepest respect ✊🏽 for all of you guys! Congratulations! 👏🏽👏🏽👏🏽 #DeepMind #ProteinStructurePrediction
Congratulations to @demishassabis and John Jumper who have won the 2023 Breakthrough Prize in Life Sciences for the development of #AlphaFold, our AI system that solved the 50-year-old challenge of protein structure prediction. 1/
Threading is fine for #ProteinStructurePrediction if you have a model. A new method for the rest is very welcome
A new method to predict the 3D structures of proteins – latest #eLIFEdigest on Medium medium.com/lifes-building…
‘It will change everything’: DeepMind’s AI makes gigantic leap in solving protein structures #ProteinStructurePrediction #BqCBemeUV nature.com/articles/d4158…
#ProteinStructurePrediction 👇 Banking on protein structural data science.sciencemag.org/content/373/65…
Microsoft が BioEmu-1 をリリース: タンパク質構造予測のためのディープラーニング モデル - InfoQ #ArchitecturalInnovation #SoftwareDevelopment #ProteinStructurePrediction #TechLeadership prompthub.info/98573/
prompthub.info
Microsoft が BioEmu-1 をリリース: タンパク質構造予測のためのディープラーニング モデル – InfoQ - プロンプトハブ
QCon San Francisco(11月17日〜21日):主要な実践者がどのように考え、複雑なソフトウェア
Take a look at this wonderfully comprehensive video from @QuantaMagazine — shows the significance of advances in #computationalproteindesign and #proteinstructureprediction, as recognized with the 2024 Nobel Prize in Chemistry! youtube.com/watch?v=cx7l9Z…
youtube.com
YouTube
How AI Cracked the Protein Folding Code and Won a Nobel Prize
The #NobelPrize for #ProteinStructurePrediction highlights the importance of protein folding and #bioAI. We are working on OpenFold3, an open-source, AlphaFold3-inspired technology. Email info-at-openfold.io to join our team! nytimes.com/2024/10/09/sci…
BREAKING NEWS The Royal Swedish Academy of Sciences has decided to award the 2024 #NobelPrize in Chemistry with one half to David Baker “for computational protein design” and the other half jointly to Demis Hassabis and John M. Jumper “for protein structure prediction.”

A second 2024 @NobelPrize involving #AI, this time in Chemistry for contributions in the field of #proteinstructureprediction, most notably with @demishassabis at @GoogleDeepMind and their groundbreaking work with #AlphaFold 🤖🧪🧬🦠🔬 #NobelPrizeChemistry
BREAKING NEWS The Royal Swedish Academy of Sciences has decided to award the 2024 #NobelPrize in Chemistry with one half to David Baker “for computational protein design” and the other half jointly to Demis Hassabis and John M. Jumper “for protein structure prediction.”

AI で科学研究を加速: Anna Koivunieme 氏へのインタビュー | McKinsey #AIinnovation #responsibilityandsafety #proteinstructureprediction #futureofAI prompthub.info/53768/
prompthub.info
AI で科学研究を加速: Anna Koivunieme 氏へのインタビュー | McKinsey - プロンプトハブ
1950年代後半以来、スーパーコンピューターの支援を受けても、人間は約20万種類のタンパク質の構造を予測できる
Revolutionizing membrane protein design with AI-powered secondary structure prediction. New tools empower the field. #MembraneProteinDesign #ProteinStructurePrediction #AIProteinDesign Details:spj.science.org/doi/10.34133/2…

Using both X-ray and microED data, the structure of the main fragment of a protein of unkown function was solved using a predicted model of that domain as a starting point @IUCr #ElectronDiffraction #ProteinStructurePrediction #BacterialProteins doi.org/10.1107/S20597…

Interesting paper alert! Learn about how AlphaFold2 predicts conditionally folding regions in intrinsically disordered proteins. 🔗Check it out here: mdpi.com/2093342.. #proteinstructureprediction #AlphaFold2 #research

Justin E. Miller et al.: AlphaFold-assisted structure determination of a bacterial protein of unknown function using X-ray and electron crystallography #BacterialProteins #ProteinStructurePrediction @ucla... #IUCr scripts.iucr.org/cgi-bin/paper?…
"Exciting breakthrough in protein structure prediction! This new AI method can handle diverse biologically relevant molecules. #AI #MachineLearning #ProteinStructurePrediction towardsai.net/p/machine-lear…"
🥳Congratulations to @DemisHassabis and John Jumper @DeepMind for winning the 2023 #LaskerAward for their work on computational #ProteinStructurePrediction with #AlphaFold #Lasker2023 #LaskerLaureate
Revolutionizing membrane protein design with AI-powered secondary structure prediction. New tools empower the field. #MembraneProteinDesign #ProteinStructurePrediction #AIProteinDesign Details:spj.science.org/doi/10.34133/2…

In Silico Structural Evaluation of Short Cationic Antimicrobial Peptides sci.fo/513 #MolecularDynamics #ProteinStructurePrediction @MDPIpharma

"The major leap in structure prediction that AlphaFold has spawned does not mean that experimental techniques have suddenly become meaningless" #AlphaFold #ProteinStructurePrediction #Crystallography doi.org/10.1107/S20532…

Meta’s generative AI ESMFold uses a large language model to predict protein structures from sequences. With lightning-fast predictions of over 700 million proteins, it's revolutionizing biotech. #AIinBiotech #ProteinStructurePrediction

Using both X-ray and microED data, the structure of the main fragment of a protein of unkown function was solved using a predicted model of that domain as a starting point @IUCr #ElectronDiffraction #ProteinStructurePrediction #BacterialProteins doi.org/10.1107/S20597…

Accurate prediction of protein structures and interactions using a 3-track network biorxiv.org/content/10.110… // #DeepMind #ProteinStructurePrediction

Interesting paper alert! Learn about how AlphaFold2 predicts conditionally folding regions in intrinsically disordered proteins. 🔗Check it out here: mdpi.com/2093342.. #proteinstructureprediction #AlphaFold2 #research

AlphaFold @ CASP13: “What just happened?” | Some Thoughts on a Mysterious Universe Blog moalquraishi.wordpress.com/2018/12/09/alp… #ProteinStructurePrediction



Just submitted last target for #CASP. Bring on the results in December! That makes a total of 426 ab initio models with our #proteinstructureprediction pipeline RBO-Aleph, #contactpredictions for 90 targets w. Epsilon-CP & 85 XL data-assisted #models robotics.tu-berlin.de/menue/research…


Something went wrong.
Something went wrong.
United States Trends
- 1. #UFC321 53.8K posts
- 2. Ole Miss 10.3K posts
- 3. Oklahoma 14.9K posts
- 4. Arbuckle 1,648 posts
- 5. Lane Kiffin 5,026 posts
- 6. UCLA 6,939 posts
- 7. Hugh Freeze 1,059 posts
- 8. Jackson Arnold N/A
- 9. Cunha 69.8K posts
- 10. Anthony Taylor 11.6K posts
- 11. Lamar 17.9K posts
- 12. #Huskers N/A
- 13. #MUFC 28.1K posts
- 14. Almeida 37.5K posts
- 15. Sunderland 118K posts
- 16. Cam Coleman 1,166 posts
- 17. Mateer 1,428 posts
- 18. #iufb 1,950 posts
- 19. Sark 3,104 posts
- 20. Northwestern 4,981 posts