#bioinformaitcs search results
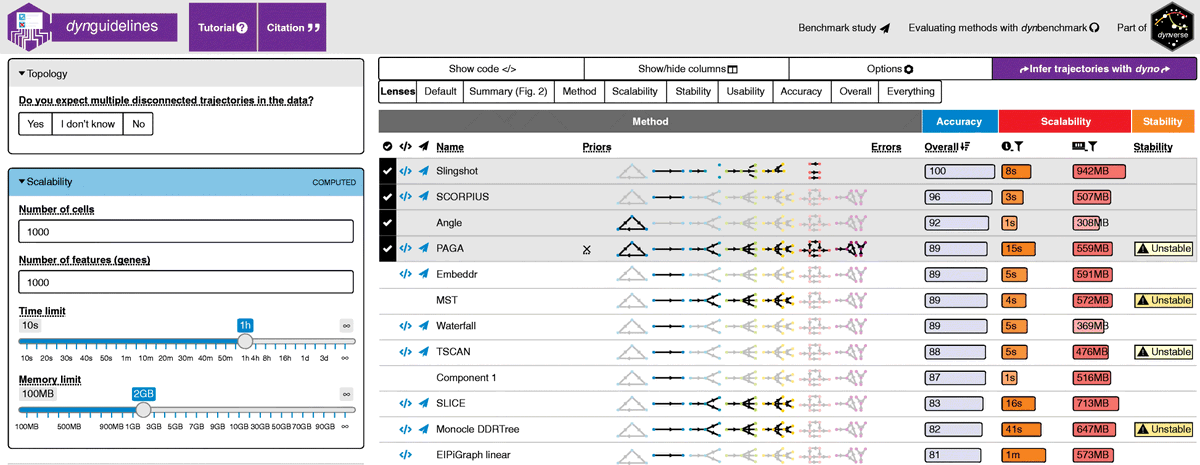
Essential guidelines for computational method benchmarking | Genome Biology genomebiology.biomedcentral.com/articles/10.11… #bioinformaitcs


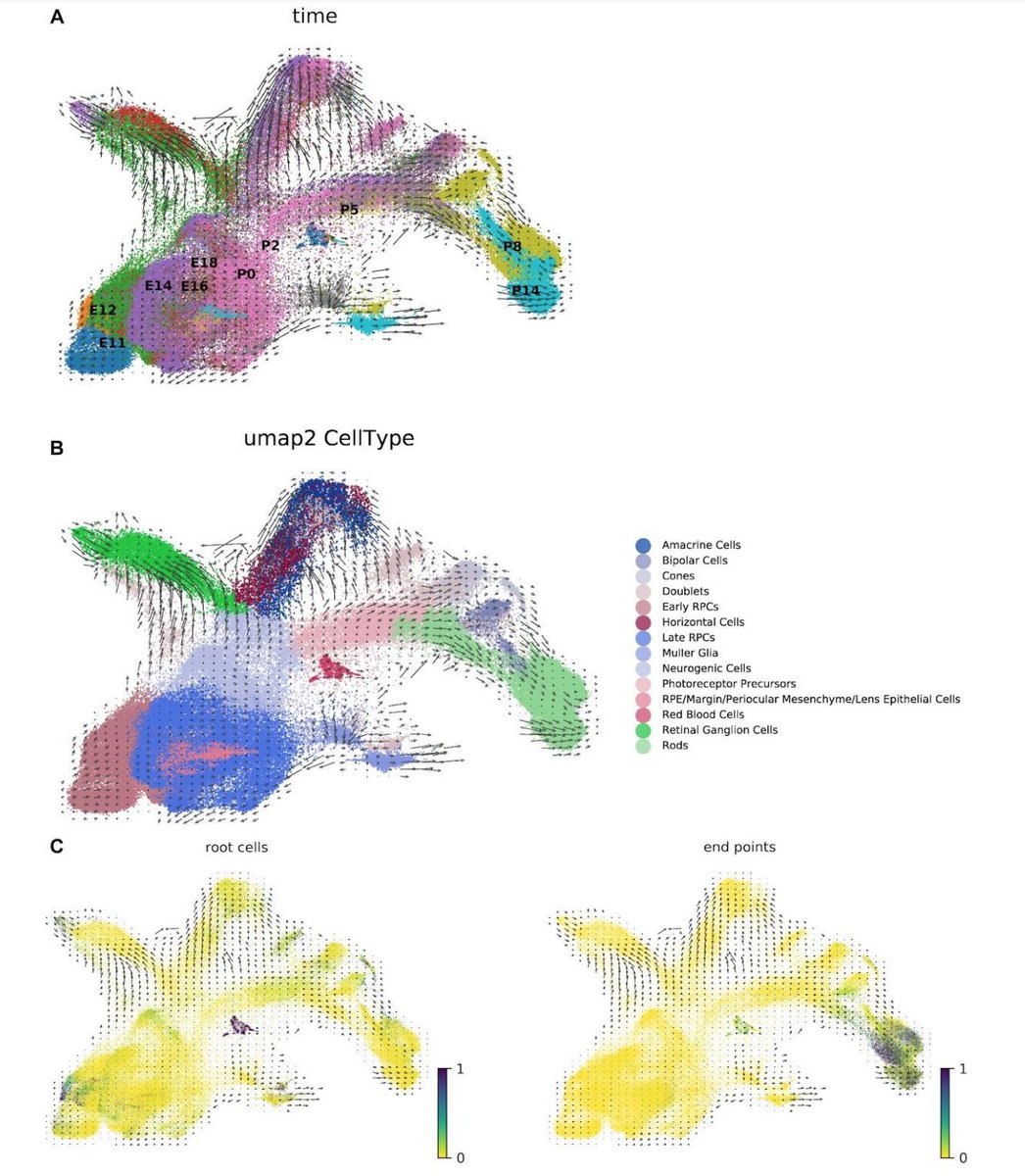
Modular and efficient pre-processing of single-cell RNA-seq | bioRxiv biorxiv.org/content/10.110… #bioinformaitcs #RNAseq


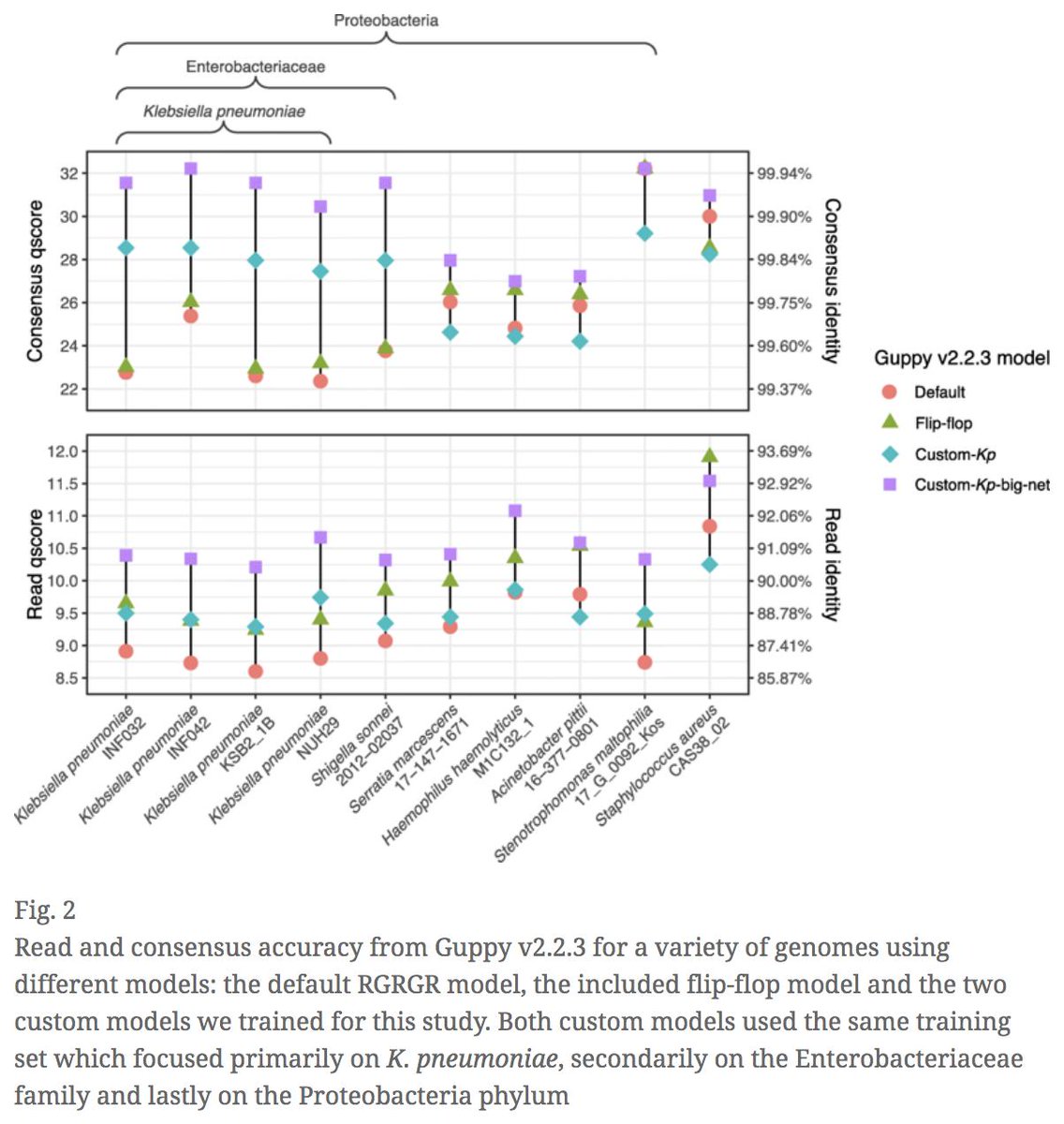
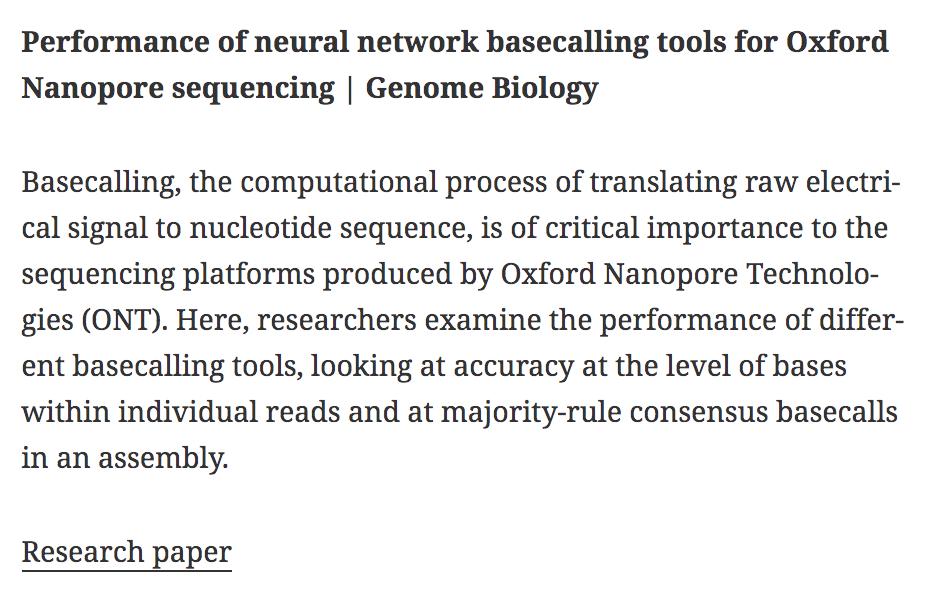
Performance of neural network basecalling tools for Oxford Nanopore sequencing | Genome Biology genomebiology.biomedcentral.com/articles/10.11… #bioinformaitcs



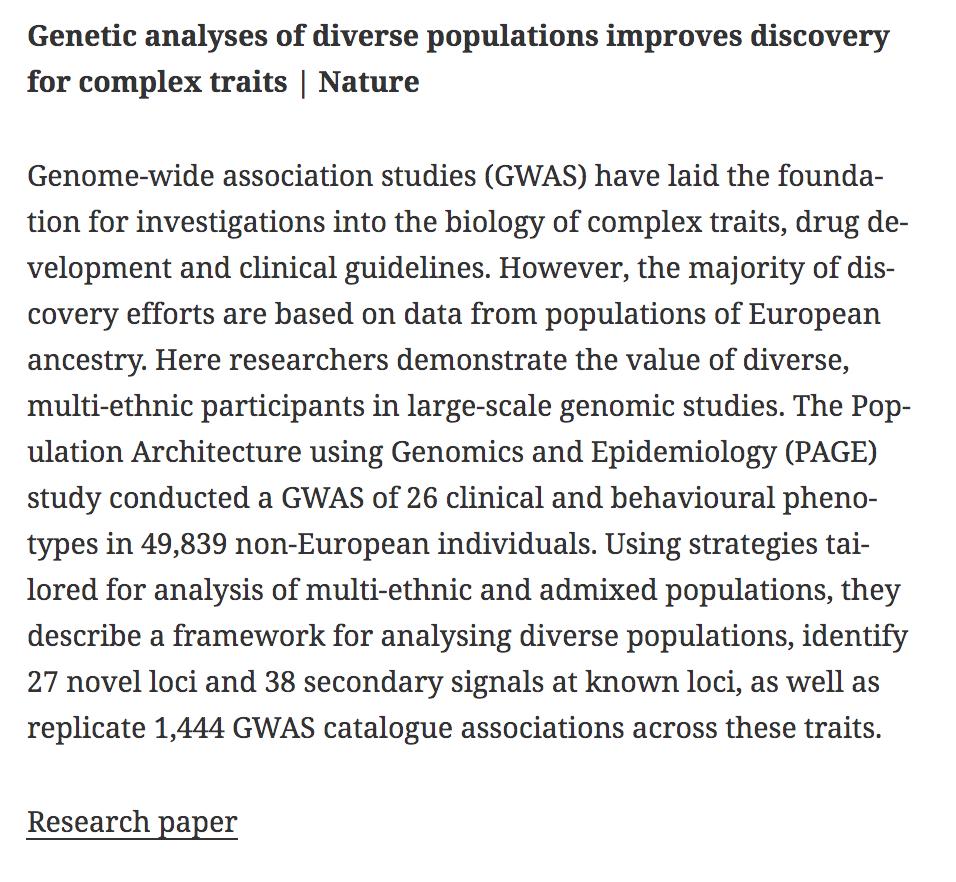
BioDecoded Daily Digest | June 22, 2019 biodecoded.com/2019/06/22/dai… #bioinformaitcs #GWAS #HumanCellAtlas


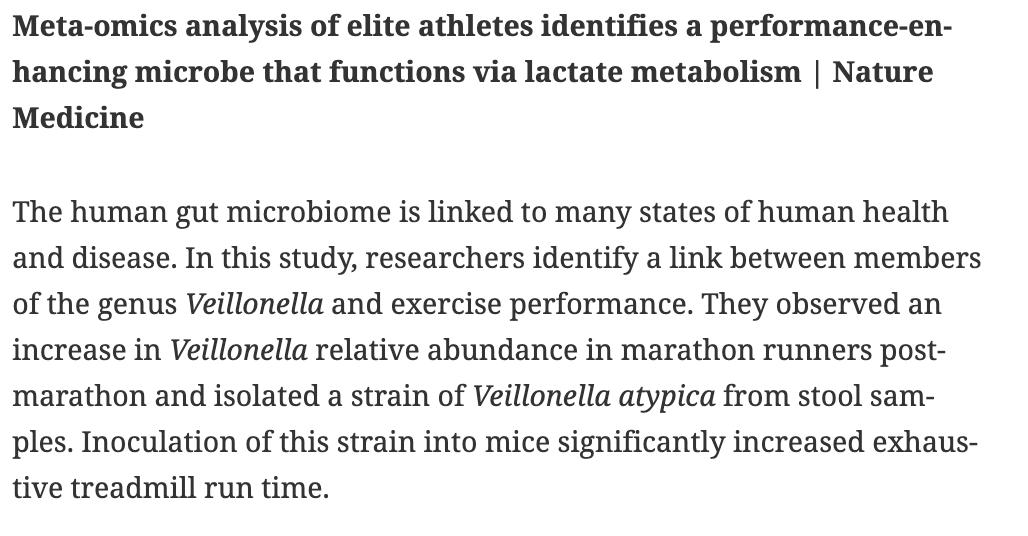
BioDecoded Daily Digest | June 24, 2019 biodecoded.com/2019/06/24/dai… #exercise #bioinformaitcs #CancerResearch
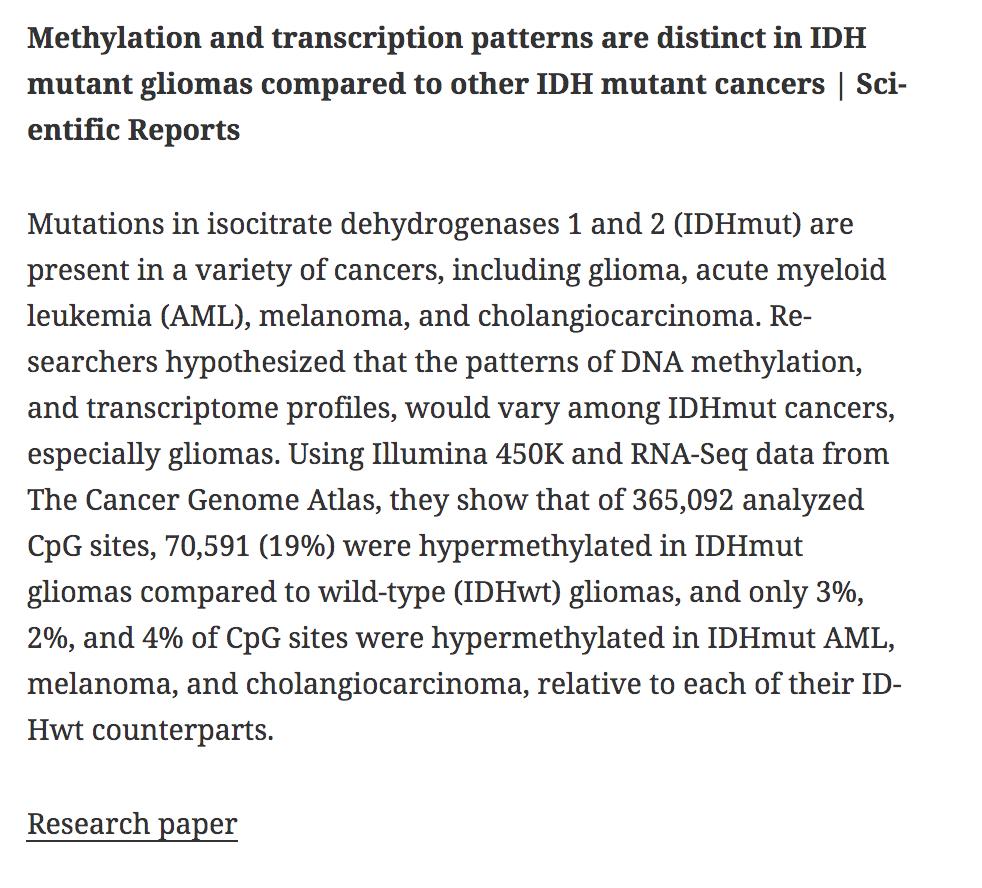
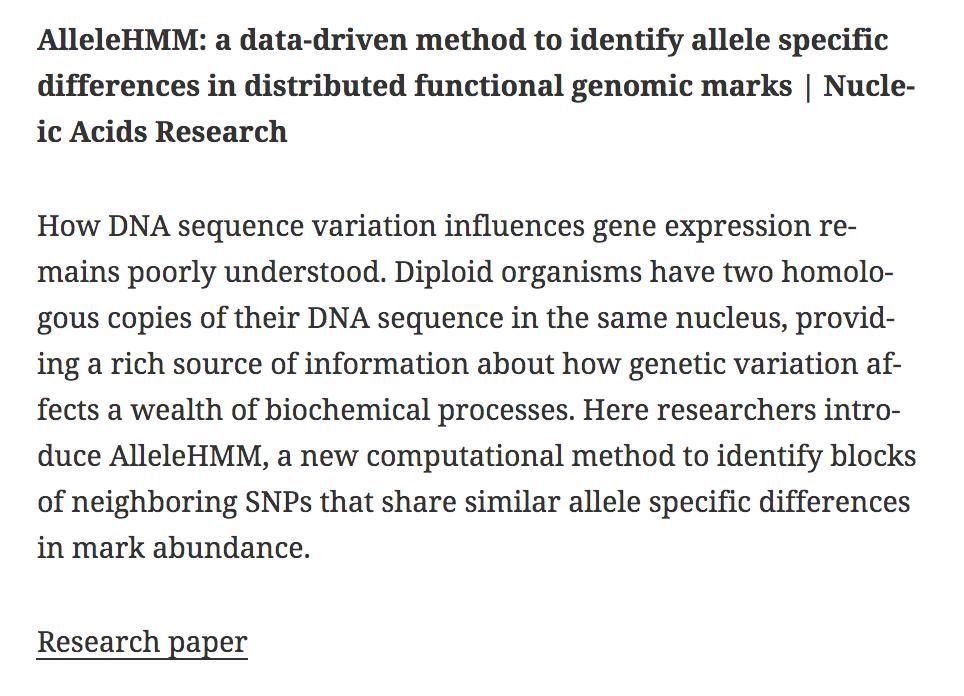

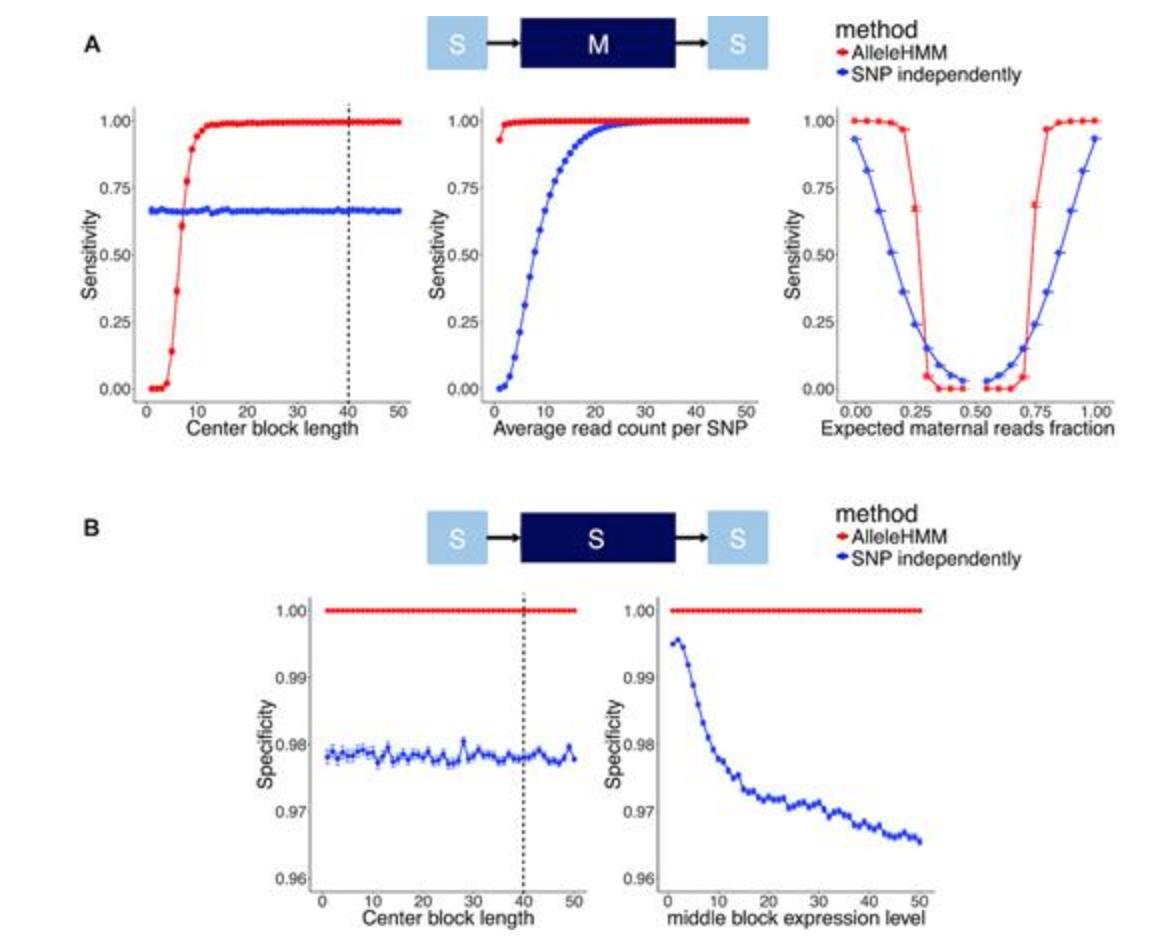
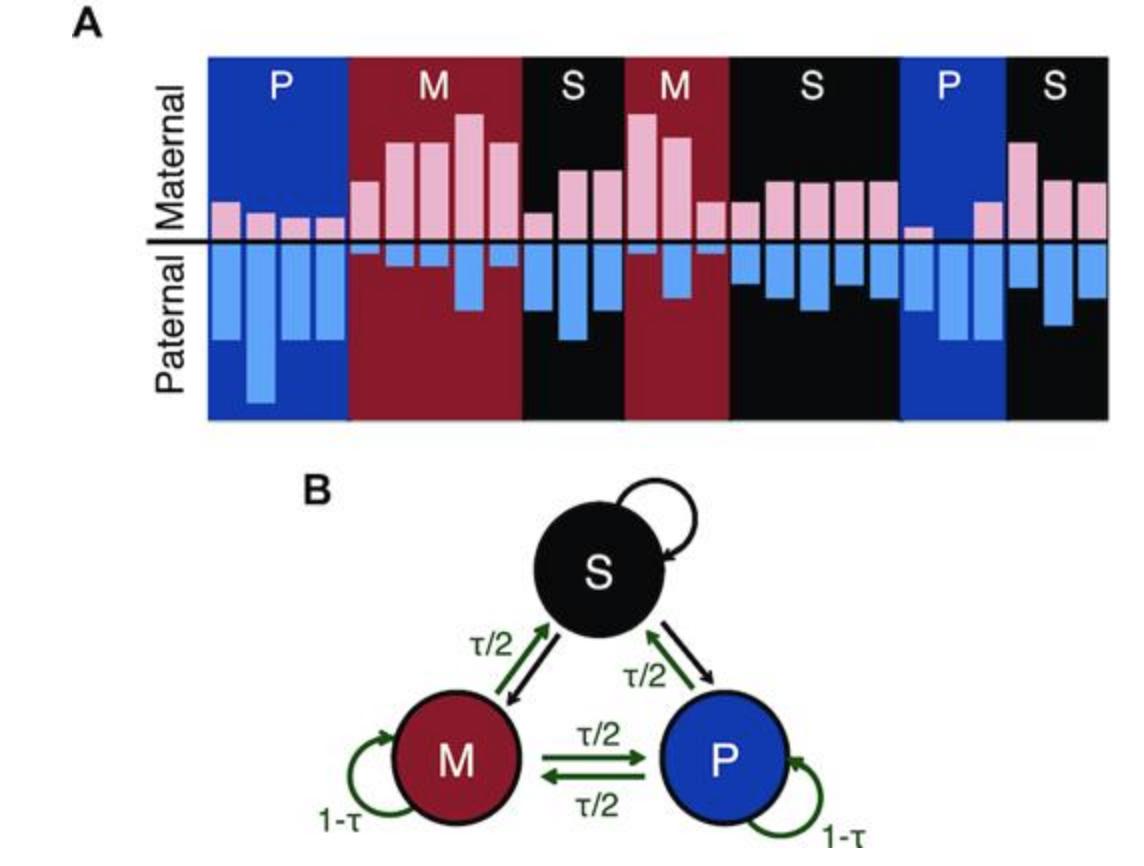
AlleleHMM: a data-driven method to identify allele specific differences in distributed functional genomic marks | Nucleic Acids Research academic.oup.com/nar/article/47… #bioinformaitcs


We have drastically reduced the amount of bandwidth required to load GAIA fully. It should load faster and be more accessible now. Try it out at bar.utoronto.ca/gaia/ #Science #Bioinformaitcs
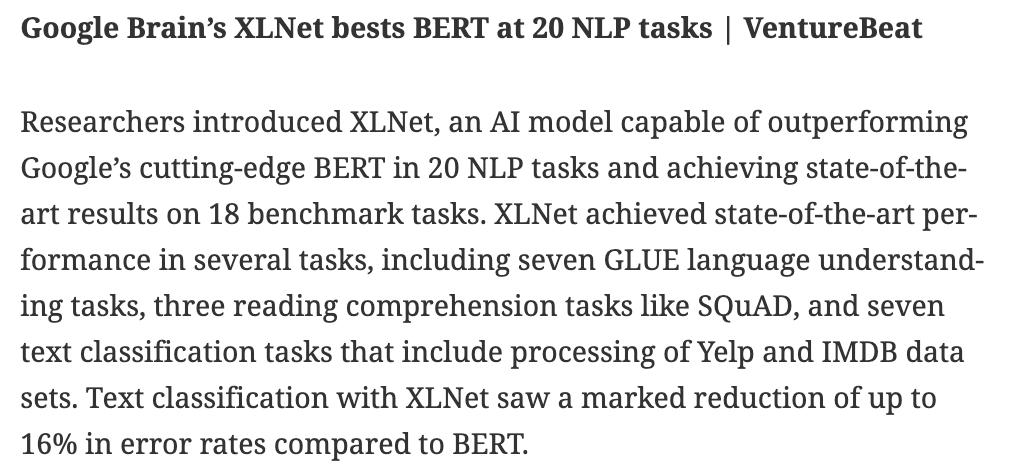
BioDecoded Daily Digest | June 26, 2019 biodecoded.com/2019/06/26/dai… #genomics #bioinformaitcs #MachineLearning #CodeSearch


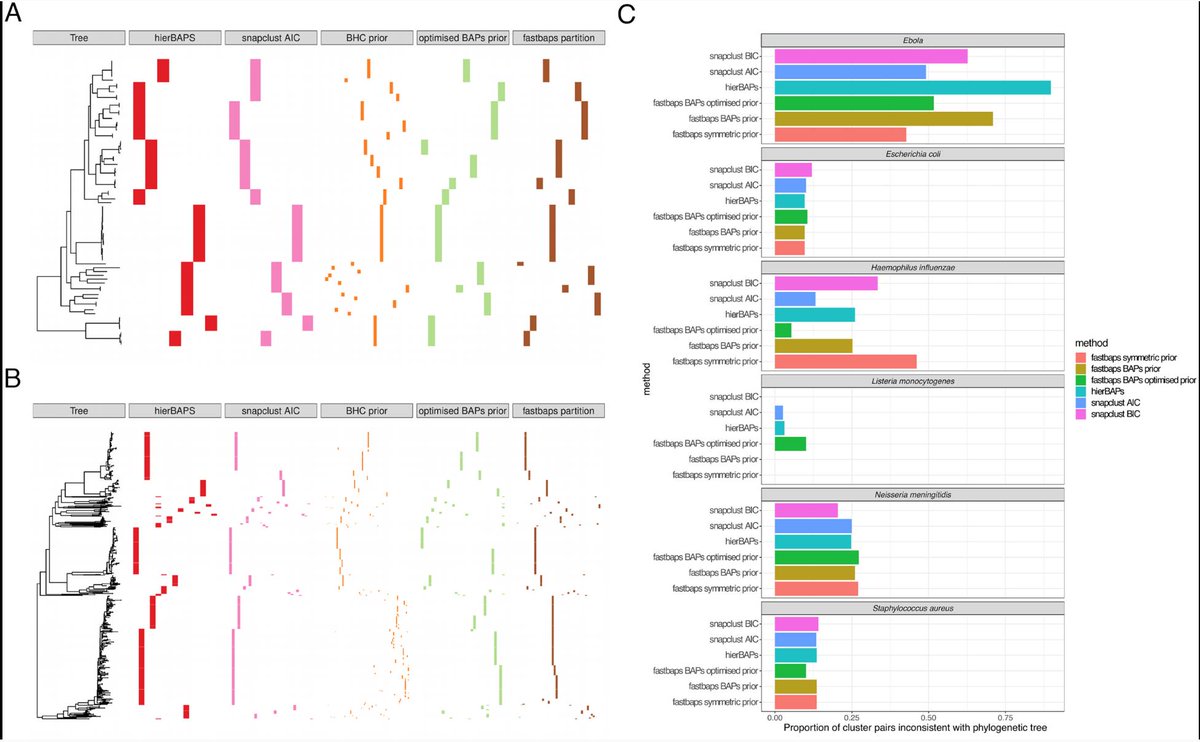
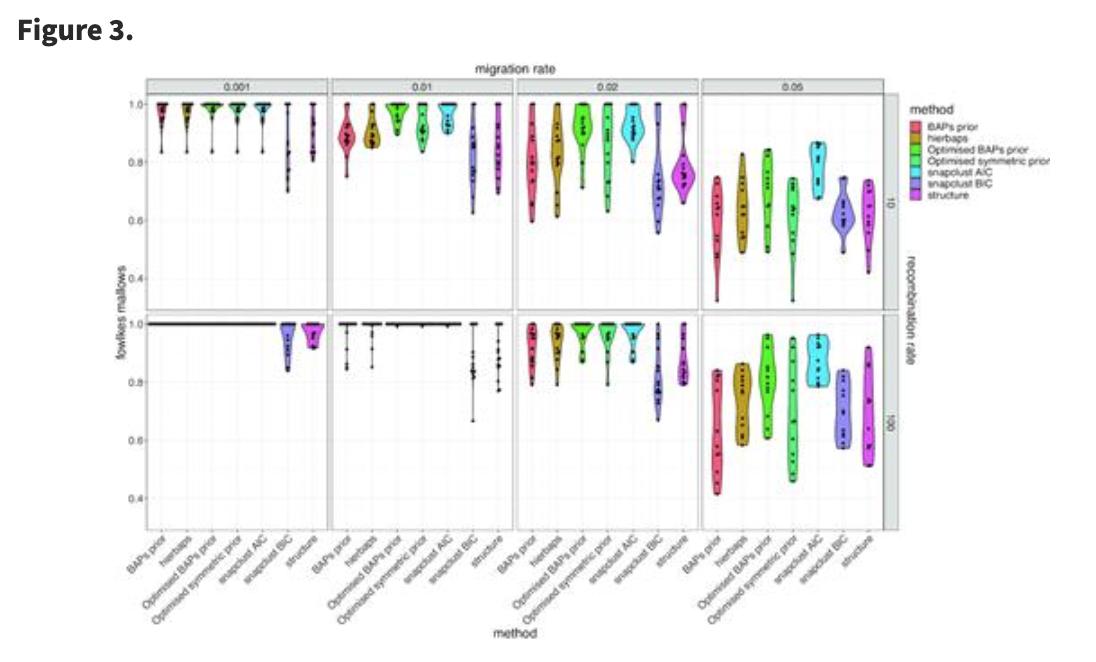
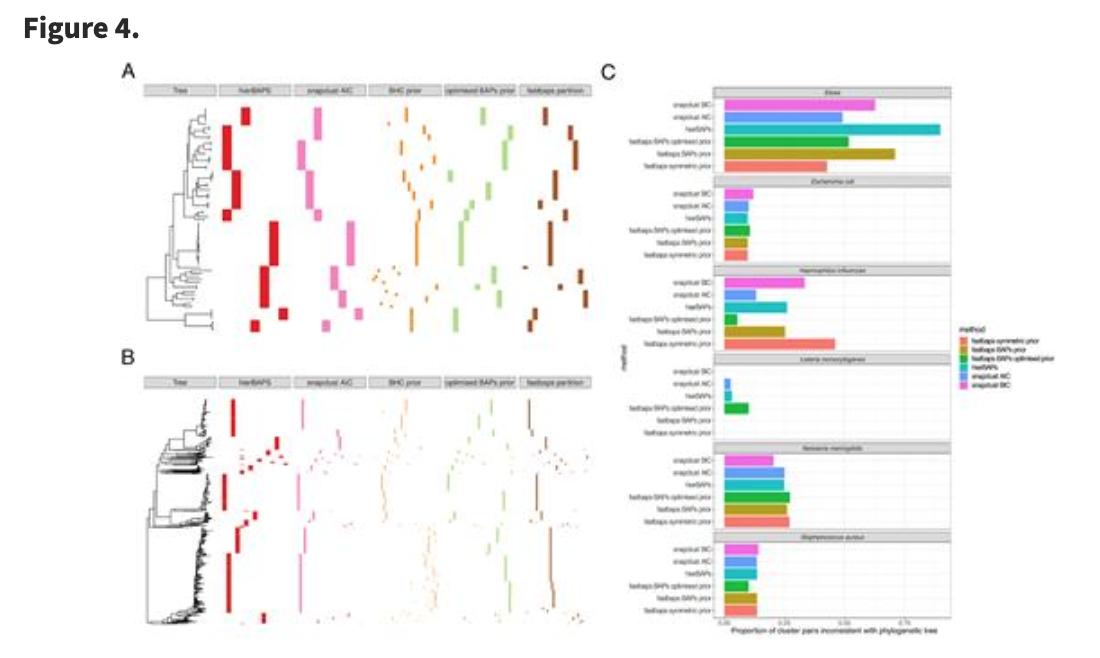
Fast hierarchical Bayesian analysis of population structure | Nucleic Acids Research academic.oup.com/nar/article/47… github.com/gtonkinhill/fa… #bioinformaitcs



BioDecoded Daily Digest | June 28, 2019 biodecoded.com/2019/06/28/dai… #CancerResearch #DecisionAnalysis #bioinformaitcs


BioDecoded Daily Digest | June 23, 2019 biodecoded.com/2019/06/23/dai… #DNA #microscopy #bioinformaitcs #RNAseq


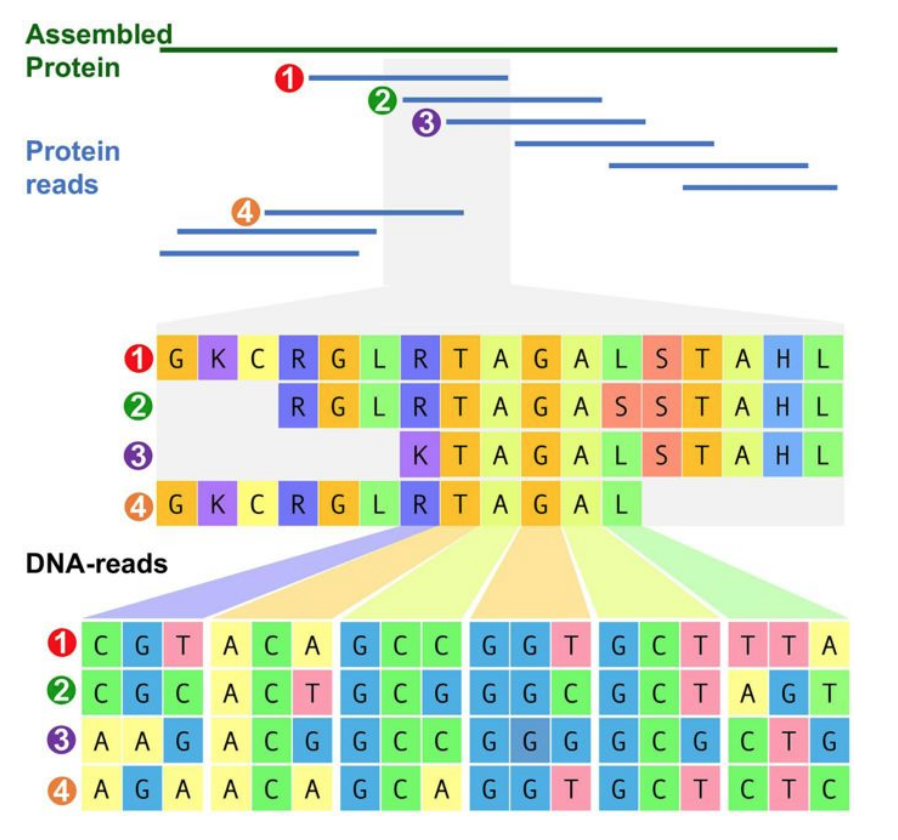
Protein-level assembly increases protein sequence recovery from metagenomic samples manyfold | Nature Methods nature.com/articles/s4159… #bioinformaitcs
BioDecoded Daily Digest | June 27, 2019 biodecoded.com/2019/06/27/dai… #bioinformaitcs #protein #music #AI #lncRNA


BioDecoded Daily Digest | June 13, 2019 biodecoded.com/2019/06/13/dai… #metagenomics #bioinformaitcs #InternetTrends2019



BioDecoded Daily Digest | August 25, 2019 biodecoded.com/2019/08/25/dai… #GutMicrobiome #bioinformaitcs #DeepLearning #NLP

Gene Information eXtension (GIX): effortless retrieval of gene product information on any website | Nature Methods nature.com/articles/s4159… github.com/knightjdr/gene… #bioinformaitcs

Bioinformatics one-liner day 10: sort VCF with header ``` cat my.vcf | awk '$0~"^#" { print $0; next } { print $0 | "sort -k1,1V -k2,2n" }' ``` #bioinformaitcs #oneliner #unix
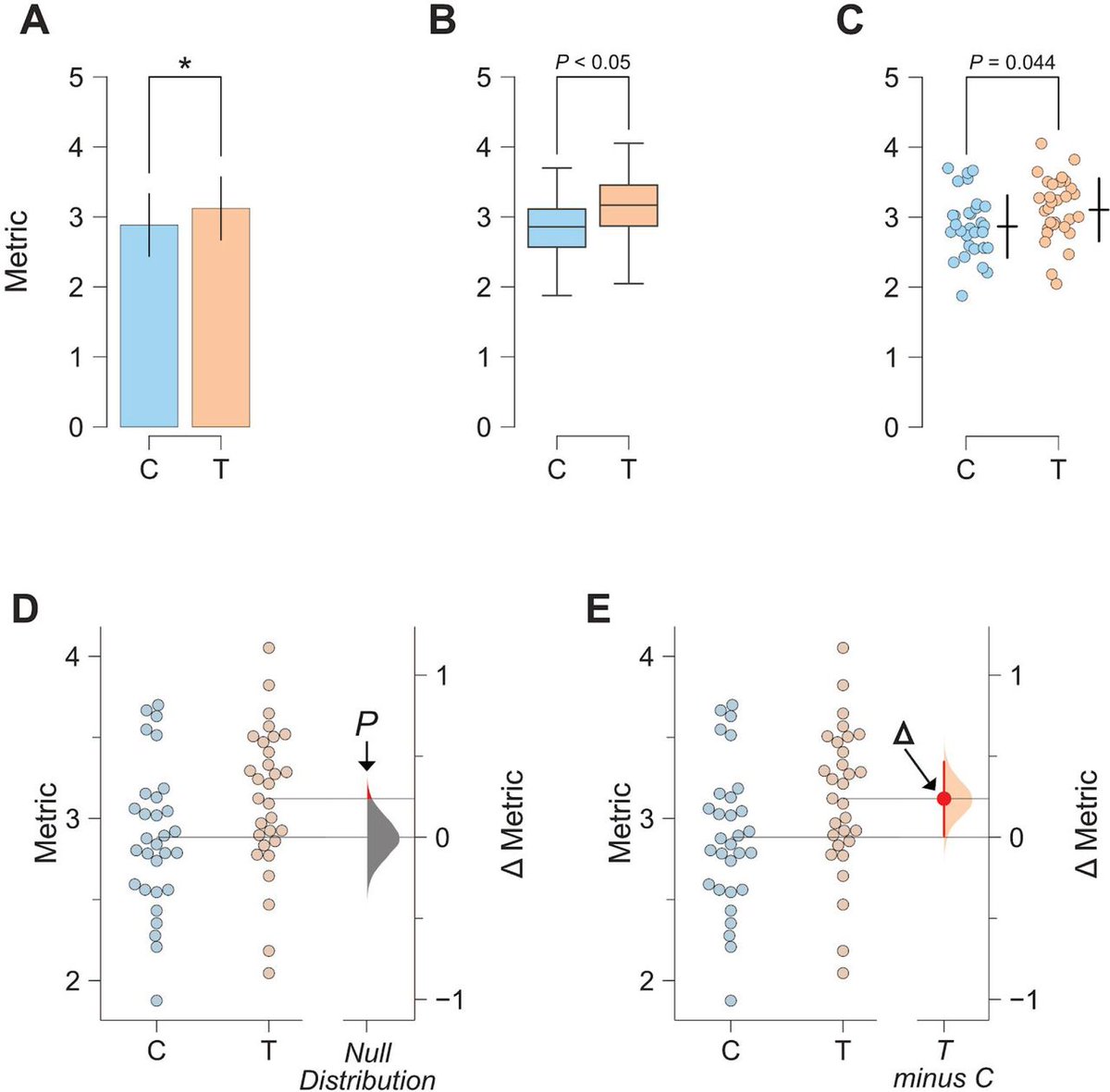
Moving beyond P values: data analysis with estimation graphics | Nature Methods nature.com/articles/s4159… biorxiv.org/content/10.110… #bioinformaitcs


Genotype Imputation from Large Reference Panels | Annual Review of Genomics and Human Genetics annualreviews.org/doi/abs/10.114… #bioinformaitcs
MYB68 regulates suberin patterning and radially distinct endodermal differentiation doi.org/10.1101/2024.0… Automatic identification and annotation of MYB gene family members in plants doi.org/10.1101/2021.1… #Bioinformaitcs #BigData
MYB68 regulates suberin patterning and radially distinct endodermal differentiation doi.org/10.1101/2024.0… Automatic identification and annotation of MYB gene family members in plants doi.org/10.1101/2021.1… #Bioinformaitcs #BigData
Bioinformatics one-liner day 10: sort VCF with header ``` cat my.vcf | awk '$0~"^#" { print $0; next } { print $0 | "sort -k1,1V -k2,2n" }' ``` #bioinformaitcs #oneliner #unix
We have drastically reduced the amount of bandwidth required to load GAIA fully. It should load faster and be more accessible now. Try it out at bar.utoronto.ca/gaia/ #Science #Bioinformaitcs

Hi #Bioinformaitcs twitter: What is your favorite aligner for genome-wide alignment of two or more species? Has anyone tried minimap2 for pairwise genomic alignments?
August 25, 2019 #DeepLearning #GutMicrobiome #bioinformaitcs via twinybots.ch biodecoded.com/2019/08/25/dai…
Genotype Imputation from Large Reference Panels | Annual Review of Genomics and Human Genetics annualreviews.org/doi/abs/10.114… #bioinformaitcs
BioDecoded Daily Digest | August 25, 2019 biodecoded.com/2019/08/25/dai… #GutMicrobiome #bioinformaitcs #DeepLearning #NLP



Fast hierarchical Bayesian analysis of population structure | Nucleic Acids Research academic.oup.com/nar/article/47… github.com/gtonkinhill/fa… #bioinformaitcs

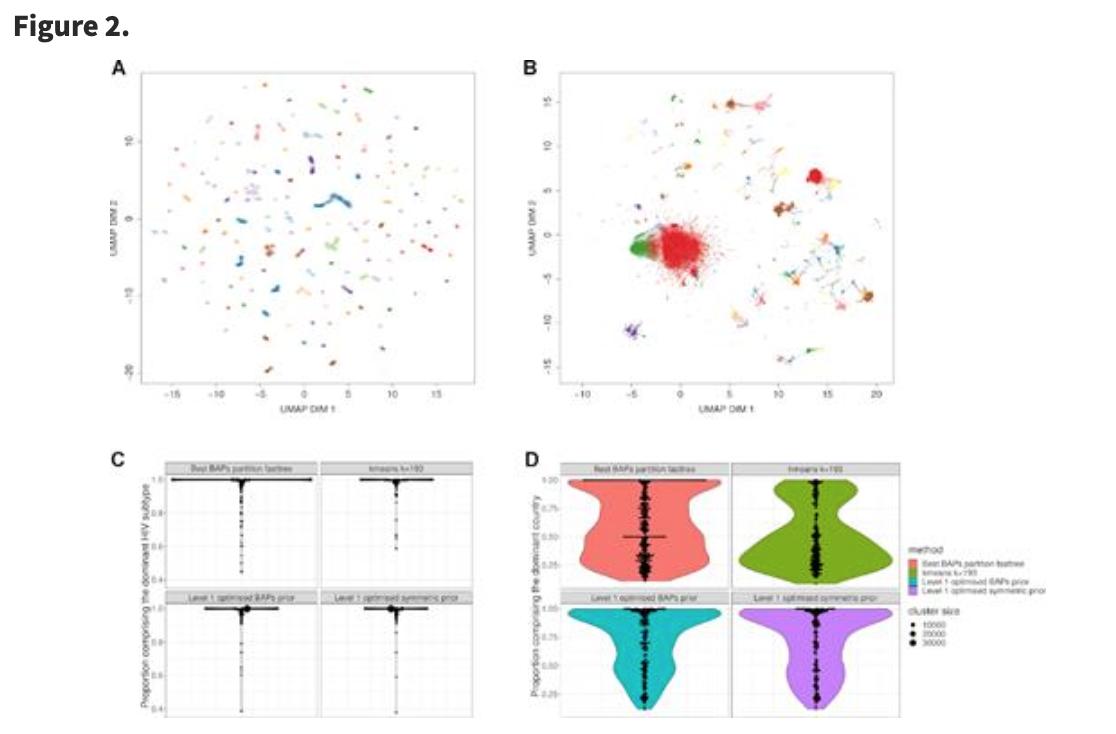
BioDecoded Daily Digest | June 28, 2019 biodecoded.com/2019/06/28/dai… #CancerResearch #DecisionAnalysis #bioinformaitcs



BioDecoded Daily Digest | June 27, 2019 biodecoded.com/2019/06/27/dai… #bioinformaitcs #protein #music #AI #lncRNA



Protein-level assembly increases protein sequence recovery from metagenomic samples manyfold | Nature Methods nature.com/articles/s4159… #bioinformaitcs

BioDecoded Daily Digest | June 26, 2019 biodecoded.com/2019/06/26/dai… #genomics #bioinformaitcs #MachineLearning #CodeSearch



Performance of neural network basecalling tools for Oxford Nanopore sequencing | Genome Biology genomebiology.biomedcentral.com/articles/10.11… #bioinformaitcs




BioDecoded Daily Digest | June 25, 2019 biodecoded.com/2019/06/25/dai… #GutMicrobiome #bioinformaitcs #DeepLearning #NLP



Fast hierarchical Bayesian analysis of population structure | Nucleic Acids Research academic.oup.com/nar/article/47… github.com/gtonkinhill/fa… #bioinformaitcs




BioDecoded Daily Digest | June 24, 2019 biodecoded.com/2019/06/24/dai… #exercise #bioinformaitcs #CancerResearch



AlleleHMM: a data-driven method to identify allele specific differences in distributed functional genomic marks | Nucleic Acids Research academic.oup.com/nar/article/47… #bioinformaitcs




BioDecoded Daily Digest | June 23, 2019 biodecoded.com/2019/06/23/dai… #DNA #microscopy #bioinformaitcs #RNAseq



Modular and efficient pre-processing of single-cell RNA-seq | bioRxiv biorxiv.org/content/10.110… #bioinformaitcs #RNAseq



Gene Information eXtension (GIX): effortless retrieval of gene product information on any website | Nature Methods nature.com/articles/s4159… github.com/knightjdr/gene… #bioinformaitcs

Essential guidelines for computational method benchmarking | Genome Biology genomebiology.biomedcentral.com/articles/10.11… #bioinformaitcs



BioDecoded Daily Digest | June 22, 2019 biodecoded.com/2019/06/22/dai… #bioinformaitcs #GWAS #HumanCellAtlas



BioDecoded Daily Digest | June 24, 2019 biodecoded.com/2019/06/24/dai… #exercise #bioinformaitcs #CancerResearch



BioDecoded Daily Digest | June 26, 2019 biodecoded.com/2019/06/26/dai… #genomics #bioinformaitcs #MachineLearning #CodeSearch



Modular and efficient pre-processing of single-cell RNA-seq | bioRxiv biorxiv.org/content/10.110… #bioinformaitcs #RNAseq



BioDecoded Daily Digest | June 28, 2019 biodecoded.com/2019/06/28/dai… #CancerResearch #DecisionAnalysis #bioinformaitcs



BioDecoded Daily Digest | June 23, 2019 biodecoded.com/2019/06/23/dai… #DNA #microscopy #bioinformaitcs #RNAseq



Performance of neural network basecalling tools for Oxford Nanopore sequencing | Genome Biology genomebiology.biomedcentral.com/articles/10.11… #bioinformaitcs




AlleleHMM: a data-driven method to identify allele specific differences in distributed functional genomic marks | Nucleic Acids Research academic.oup.com/nar/article/47… #bioinformaitcs




BioDecoded Daily Digest | June 27, 2019 biodecoded.com/2019/06/27/dai… #bioinformaitcs #protein #music #AI #lncRNA



BioDecoded Daily Digest | June 13, 2019 biodecoded.com/2019/06/13/dai… #metagenomics #bioinformaitcs #InternetTrends2019



BioDecoded Daily Digest | August 25, 2019 biodecoded.com/2019/08/25/dai… #GutMicrobiome #bioinformaitcs #DeepLearning #NLP



Fast hierarchical Bayesian analysis of population structure | Nucleic Acids Research academic.oup.com/nar/article/47… github.com/gtonkinhill/fa… #bioinformaitcs




We have drastically reduced the amount of bandwidth required to load GAIA fully. It should load faster and be more accessible now. Try it out at bar.utoronto.ca/gaia/ #Science #Bioinformaitcs

Protein-level assembly increases protein sequence recovery from metagenomic samples manyfold | Nature Methods nature.com/articles/s4159… #bioinformaitcs

Gene Information eXtension (GIX): effortless retrieval of gene product information on any website | Nature Methods nature.com/articles/s4159… github.com/knightjdr/gene… #bioinformaitcs

Benchmarking algorithms for gene regulatory network inference from single-cell transcriptomic data | arXiv biorxiv.org/content/10.110… #bioinformaitcs




Moving beyond P values: data analysis with estimation graphics | Nature Methods nature.com/articles/s4159… biorxiv.org/content/10.110… #bioinformaitcs



Something went wrong.
Something went wrong.
United States Trends
- 1. Araujo 187K posts
- 2. Chelsea 654K posts
- 3. Barca 261K posts
- 4. Estevao 270K posts
- 5. Wizards 6,240 posts
- 6. Barcelona 466K posts
- 7. Yamal 205K posts
- 8. Hazel 9,529 posts
- 9. Ferran 79K posts
- 10. Bishop Boswell N/A
- 11. Oklahoma State 4,958 posts
- 12. Godzilla 26.3K posts
- 13. Eric Morris 3,364 posts
- 14. Skippy 5,351 posts
- 15. Leftover 6,551 posts
- 16. Witkoff 65.2K posts
- 17. Cucurella 106K posts
- 18. Rashford 24.5K posts
- 19. Raising Arizona 1,937 posts
- 20. Cooks 12.4K posts