#ms2deepscore 搜尋結果
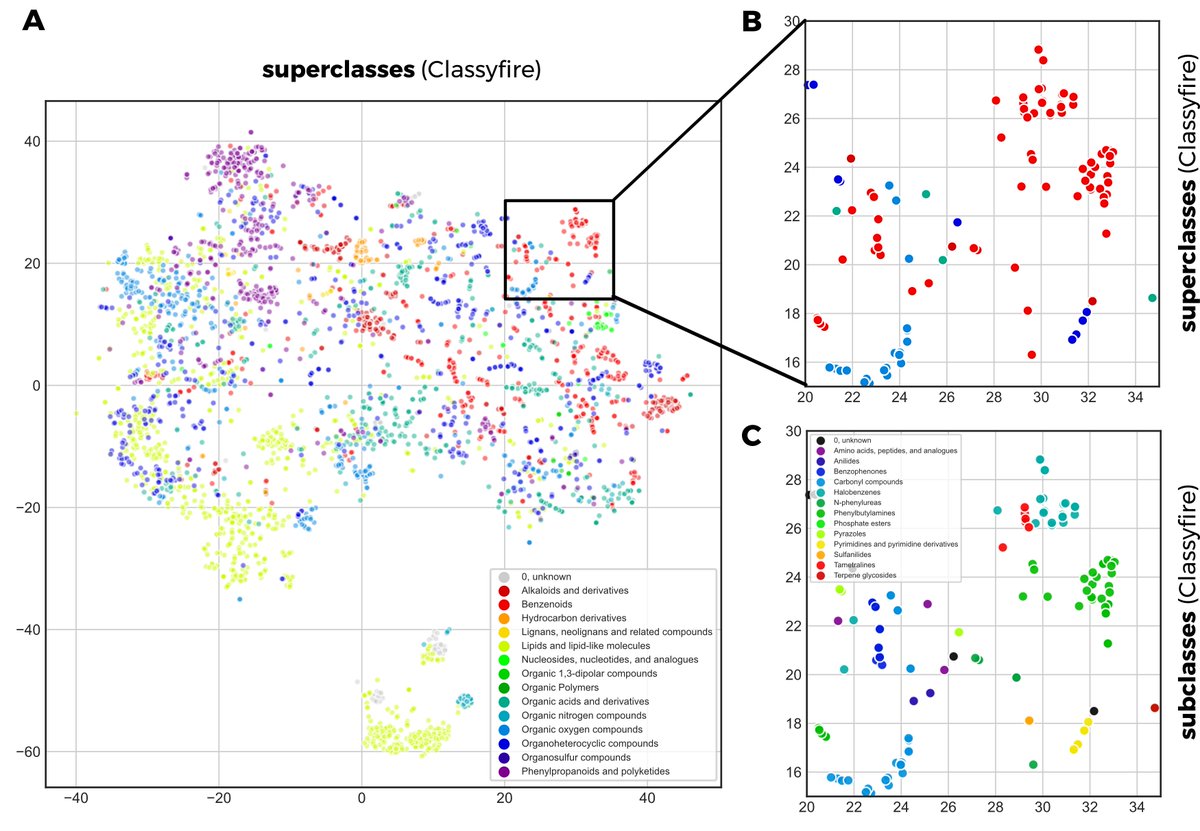
4/6 We argue that #MS2DeepScore is a promising new spectral similarity measure to complement (or replace) current metrics. We also show that it creates meaningful spectral embeddings, for instance to be used for clustering. Here on 3,600 spectra (colored by #classyfire labels).
Our newest publication is out today! 😎 #MS2DeepScore is in @jcheminf! Work led by @me_datapoint. With huge thanks to reviewers, editor, and collaborators from @eScienceCenter. @WU_BioInfo @WURplant @WUR You can read it #openacces here: jcheminf.biomedcentral.com/articles/10.11… a little thread!
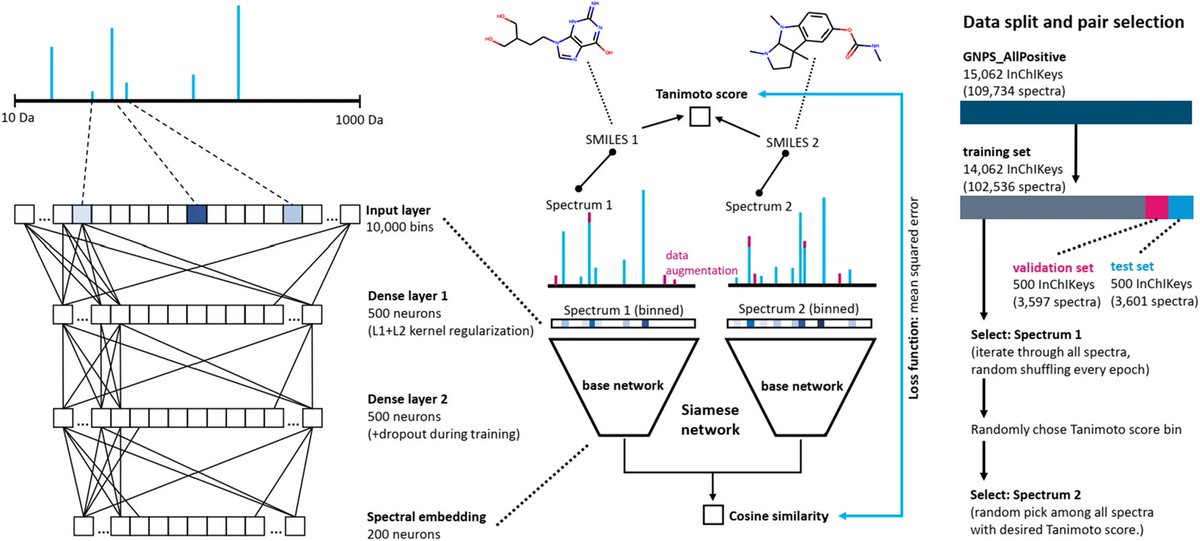
Very happy to share our work on #MS2DeepScore, a #DeepLearning approach to predict structural similarities between pairs of MS/MS spectra! A small thread on this work between @eScienceCenter/ @svenvanderburg1 and @jjjvanderhooft. biorxiv.org/content/10.110…

Excited to share #collaborative work with Florian Huber @me_datapoint, @svenvanderburg1, and Lars Ridder from the @eScienceCenter! 😎 Where our earlier #Spec2Vec is an unsupervised approach that learns mass spectral similarity, the new #MS2DeepScore is a supervised approach! 😎

Great job by Kevin Mildau who presented our recent work with @ben_warth, @enit7677, @jjjvanderhooft and others on #specXplore for explorative data analysis built on #matchms and #MS2DeepScore! 😎 #CompMetabolomics @nlmetabolomics @ynmc_benelux


The story of #MS2DeepScore started with its unsupervised cousin #Spec2Vec published earlier this year. We demonstrated how #mass #spectral #embeddings capture #chemical #information and provide novel #similarity #scores! 😎 You can read all about it here: journals.plos.org/ploscompbiol/a…
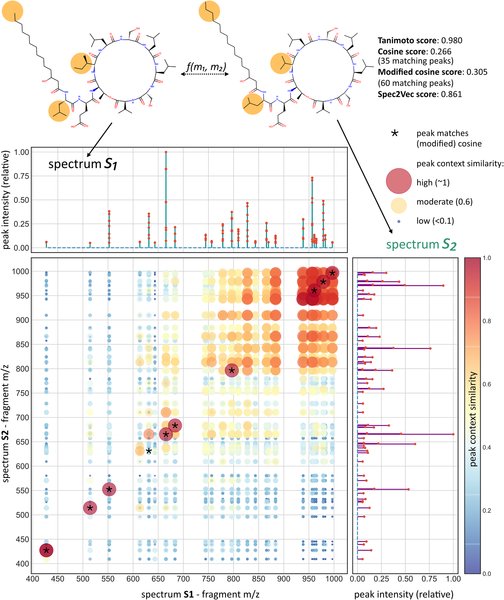
Also featuring @GNPS_UCSD and @ms2lda as well as #Spec2Vec and #MS2DeepScore! 😎
The link was broken. Congratulations 🎊 🎉 😎 👏 to all involved! 😎 Did you also try #Spec2Vec and #MS2DeepScore based similarity scores? And SiSiMe?
Congratulations 🎊 to all involved! 😎 👏 - indeed, it would be very helpful to share those MS/MS spectra with the community. Did you give @ms2lda a try to create annotated MotifSets? And with #Spec2Vec and #MS2DeepScore you could have a lot of fun with that data! @me_datapoint
Finally, it will be exciting to see how novel #MachineLearning based scores like #Spec2Vec and #MS2DeepScore, & #SIMILE will perform for these and other #analogs with >1 #chemical #modification. Yet another boost for #untargeted #massspectrometry based #metabolomics #analyses.😊
New #MS2DeepScore release (0.3.0) 🚀. --> github.com/matchms/ms2dee… Now possible to include numerical metadata entries as features (e.g. precursor m/z). #openscience #opensource #python #massspec #metabolomics #deeplearning
With @me_datapoint of course! 😎 #Spec2Vec #MS2DeepScore #embeddings #clustering #networking
With @GNPS_UCSD, @ms2lda, #matchms, #Spec2Vec, #MS2DeepScore, @antiSMASH_dev #antiSMASH #MIBiG, #BiG-SCAPE, @NatProdAtlas, #NPLinker, and many other #openscience projects, #tools, and #resources, these are exciting times for the field! 😎
Based on our #benchmarking and first explorations we argue that #MS2DeepScore is a promising new #massspectrometry #spectral #similarity #score metric to complement (or replace) current metrics for #librarymatching and #networking analysis and other possible applications! 😎
New release of #matchms (0.18.0) and other key pieces of the matchms ecosystem: #spec2vec (0.8.0) & #ms2deepscore (0.3.1).😊 --> github.com/matchms/matchms ✨ Similarity scores are stored as sparse arrays ✨ New Pipeline class to assemble matchms workflows #OpenSource #OpenScience
#MS2DeepScore is built on the #matchms #Python package that handles and processes #massspectrometry #data for #metabolomics #analyses aiming for #spectral #comparisons. The full source code is available on #GitHub: github.com/matchms/ms2dee…
Very glad to see our work on #MS2DeepScore published in @jcheminf ! This work is the result of a great collaboration with @jjjvanderhooft and @eScienceCenter (certainly not the last project we do together 😉🙌) #OpenAccess article --> jcheminf.biomedcentral.com/articles/10.11…
Great to see this one out today! 😎 Enjoy the read and have a great weekend! 😎 #MS2DeepScore #metabolomics #mass #spectral #embeddings
Our newest publication is out today! 😎 #MS2DeepScore is in @jcheminf! Work led by @me_datapoint. With huge thanks to reviewers, editor, and collaborators from @eScienceCenter. @WU_BioInfo @WURplant @WUR You can read it #openacces here: jcheminf.biomedcentral.com/articles/10.11… a little thread!

You can try out #MS2DeepScore yourself by using our #Python package --> github.com/matchms/ms2dee… Let us know what you think or if you need any help to implement our tool in your own #massspec workflow! #OpenScience
MS2Query builds on previous software tools #matchms, #Spec2Vec, and #MS2Deepscore. Working in #Python with #massspectrometry #data?! Give matchms a try! github.com/matchms/matchms
New @mzmine_project version 4.3: improved memory management, #MS2Deepscore molecular networking, fragment dashboard, MSConvert support. #metabolomics #CompMS #lipidomics Download now: github.com/mzmine/mzmine/… More information & Video: linkedin.com/posts/mzio_mzm…
Great job by Kevin Mildau who presented our recent work with @ben_warth, @enit7677, @jjjvanderhooft and others on #specXplore for explorative data analysis built on #matchms and #MS2DeepScore! 😎 #CompMetabolomics @nlmetabolomics @ynmc_benelux


We just made it in time for the Friday deadline! 💫 2 new releases: ✨#matchms 0.22.0 --> pypi.org/project/matchm… ✨#ms2deepscore 0.5.0 --> pypi.org/project/ms2dee… #opensource #openscience #massspec #Python Many thanks to Niek de Jonge & @hecht_h 🙏
New release of #matchms (0.18.0) and other key pieces of the matchms ecosystem: #spec2vec (0.8.0) & #ms2deepscore (0.3.1).😊 --> github.com/matchms/matchms ✨ Similarity scores are stored as sparse arrays ✨ New Pipeline class to assemble matchms workflows #OpenSource #OpenScience
New #MS2DeepScore release (0.3.0) 🚀. --> github.com/matchms/ms2dee… Now possible to include numerical metadata entries as features (e.g. precursor m/z). #openscience #opensource #python #massspec #metabolomics #deeplearning
MS2Query builds on previous software tools #matchms, #Spec2Vec, and #MS2Deepscore. Working in #Python with #massspectrometry #data?! Give matchms a try! github.com/matchms/matchms
Finally, it will be exciting to see how novel #MachineLearning based scores like #Spec2Vec and #MS2DeepScore, & #SIMILE will perform for these and other #analogs with >1 #chemical #modification. Yet another boost for #untargeted #massspectrometry based #metabolomics #analyses.😊
With @GNPS_UCSD, @ms2lda, #matchms, #Spec2Vec, #MS2DeepScore, @antiSMASH_dev #antiSMASH #MIBiG, #BiG-SCAPE, @NatProdAtlas, #NPLinker, and many other #openscience projects, #tools, and #resources, these are exciting times for the field! 😎
The link was broken. Congratulations 🎊 🎉 😎 👏 to all involved! 😎 Did you also try #Spec2Vec and #MS2DeepScore based similarity scores? And SiSiMe?
You can try out #MS2DeepScore yourself by using our #Python package --> github.com/matchms/ms2dee… Let us know what you think or if you need any help to implement our tool in your own #massspec workflow! #OpenScience
Our newest publication is out today! 😎 #MS2DeepScore is in @jcheminf! Work led by @me_datapoint. With huge thanks to reviewers, editor, and collaborators from @eScienceCenter. @WU_BioInfo @WURplant @WUR You can read it #openacces here: jcheminf.biomedcentral.com/articles/10.11… a little thread!

4/6 We argue that #MS2DeepScore is a promising new spectral similarity measure to complement (or replace) current metrics. We also show that it creates meaningful spectral embeddings, for instance to be used for clustering. Here on 3,600 spectra (colored by #classyfire labels).

Excited to share #collaborative work with Florian Huber @me_datapoint, @svenvanderburg1, and Lars Ridder from the @eScienceCenter! 😎 Where our earlier #Spec2Vec is an unsupervised approach that learns mass spectral similarity, the new #MS2DeepScore is a supervised approach! 😎

Great job by Kevin Mildau who presented our recent work with @ben_warth, @enit7677, @jjjvanderhooft and others on #specXplore for explorative data analysis built on #matchms and #MS2DeepScore! 😎 #CompMetabolomics @nlmetabolomics @ynmc_benelux


Very happy to share our work on #MS2DeepScore, a #DeepLearning approach to predict structural similarities between pairs of MS/MS spectra! A small thread on this work between @eScienceCenter/ @svenvanderburg1 and @jjjvanderhooft. biorxiv.org/content/10.110…

The story of #MS2DeepScore started with its unsupervised cousin #Spec2Vec published earlier this year. We demonstrated how #mass #spectral #embeddings capture #chemical #information and provide novel #similarity #scores! 😎 You can read all about it here: journals.plos.org/ploscompbiol/a…

Something went wrong.
Something went wrong.
United States Trends
- 1. National Guard 236K posts
- 2. Thanksgiving 470K posts
- 3. Arsenal 471K posts
- 4. Liverpool 151K posts
- 5. Slot 125K posts
- 6. Blood 171K posts
- 7. Bayern 245K posts
- 8. Frank Ragnow 8,336 posts
- 9. Neuer 24.7K posts
- 10. Martinelli 32K posts
- 11. Declan Rice 28.2K posts
- 12. Konate 18.5K posts
- 13. Anfield 27.4K posts
- 14. Seditious Six 121K posts
- 15. Dylan Cease 1,373 posts
- 16. Arteta 39K posts
- 17. Mbappe 124K posts
- 18. Insurrection Act 6,385 posts
- 19. #COYG 7,388 posts
- 20. Denzel 5,245 posts