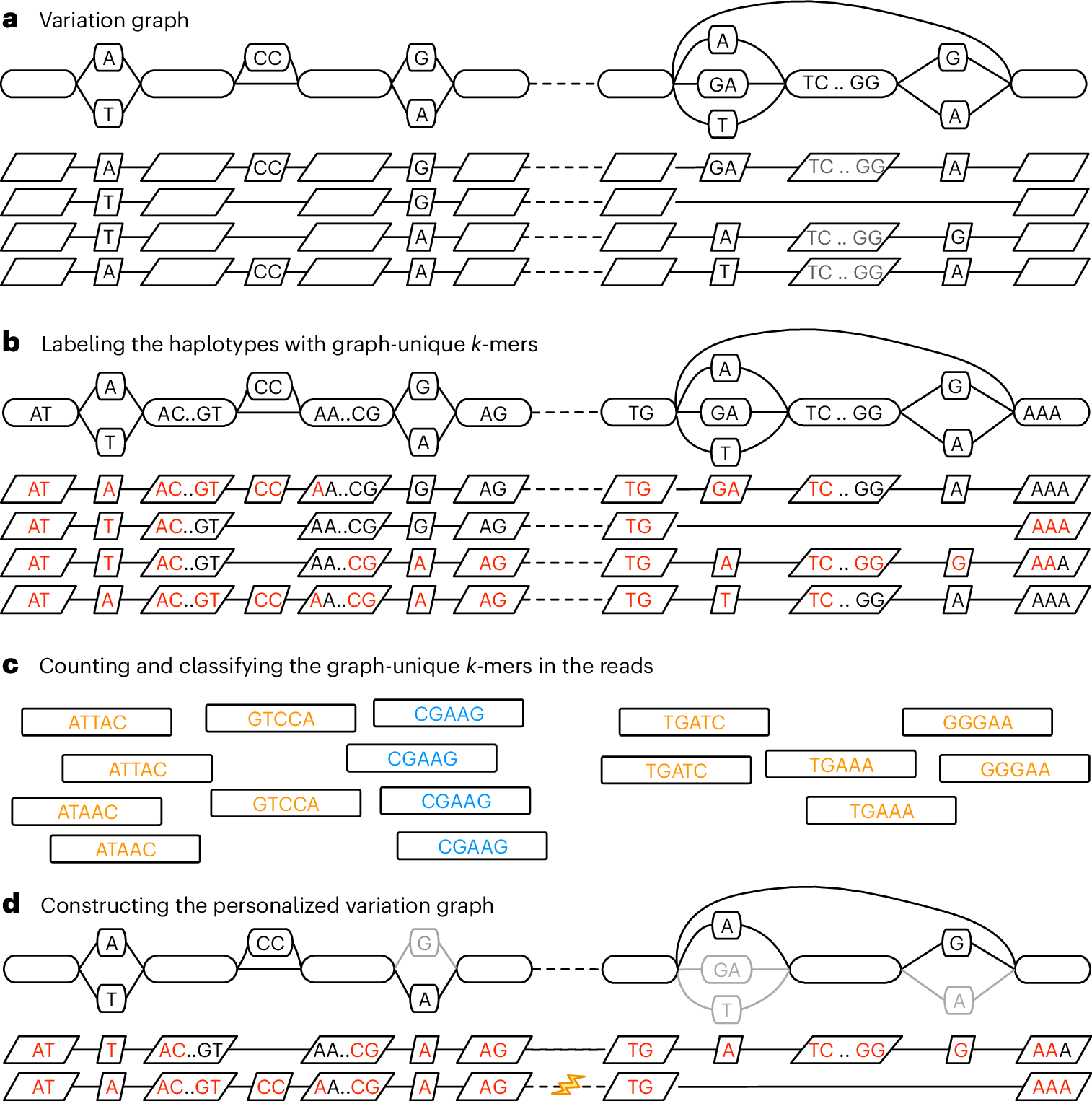
#pangenomics search results
A Pangenomic Method for Establishing a Somatic Variant Detection Resource in HapMap Mixtures. #SomaticVariantDetection #VariantCalling #Pangenomics #Genomics #Bioinformatics @biorxiv_genomic biorxiv.org/content/10.110…

An intriguing study that puts an 800-year-old Salmonella genome from a skeleton in the context of 50K modern Salmonella genomes: doi.org/10.1016/j.cub.…. Excellent use of #metagenomics & #pangenomics by Zhemin Zhou et al. Plus the most beautiful #anvio figure ever published!

Billi: Provably Accurate and Scalable Bubble Detection in Pangenome Graphs doi.org/10.1101/2025.1… Pangenome of cultivated beet and crop wild relatives reveals parental relationships of a tetraploid wild beet doi.org/10.1101/2023.0… #Genomics #Bioinformatics #Pangenomics
This Wednesday at 9am CST I will give a webinar on #pangenomics. Please fill this form for a Zoom link if you'd like to attend: forms.gle/WaUmDQzqxvFXip… Please note that this webinar will target beginners and will discuss 'basics' rather than 'practice'.

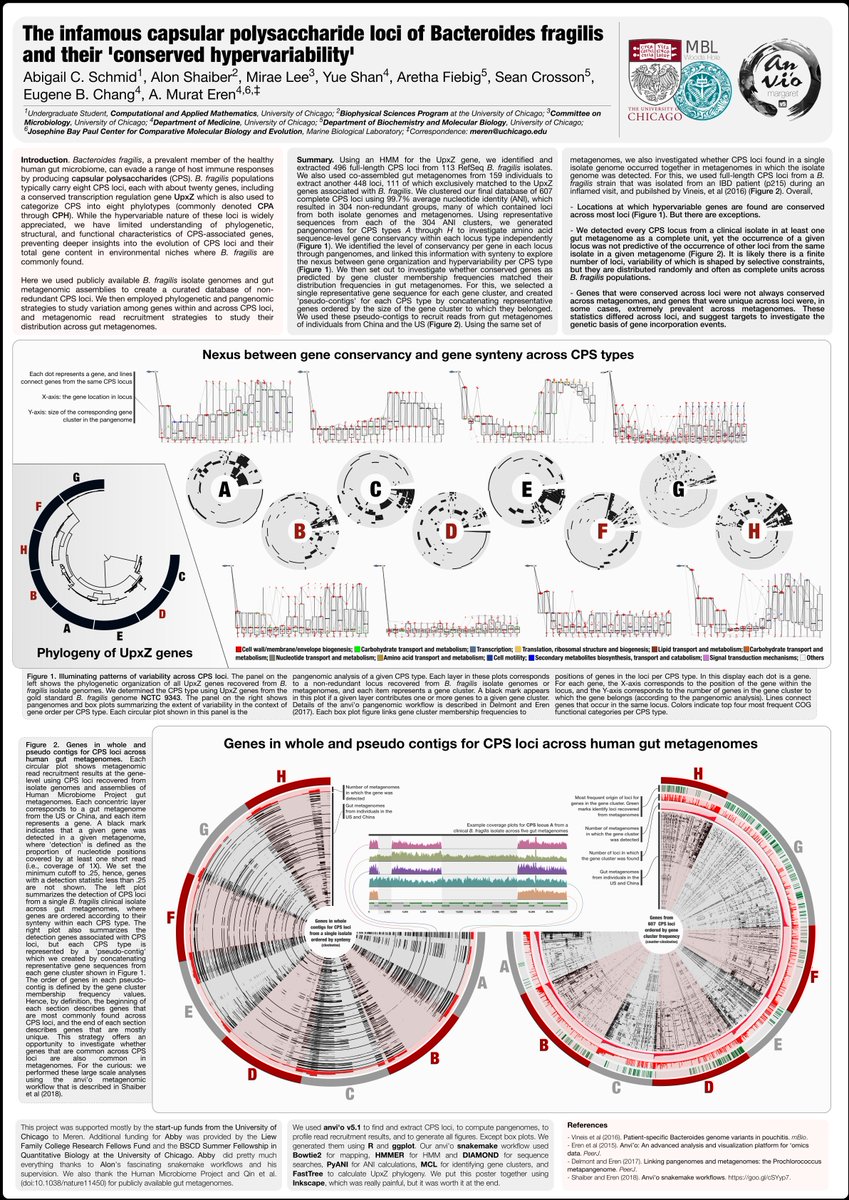
Visit poster 533A today at #ISME17 to hear from Abby Schmid about our attempt to characterize all capsular polysaccharide loci from all Bacteroides fragilis isolates in the context of many human guts using #pangenomics #metagenomics and #anvio.
Participant profile of today's webinar on #pangenomics. Participants who filled the survey asked for (1) a moderator to bring up questions from the chat window, (2) a follow-up on applications of pangenomics, & (3) a technical hands-on session. Challenge accepted. Thank you!

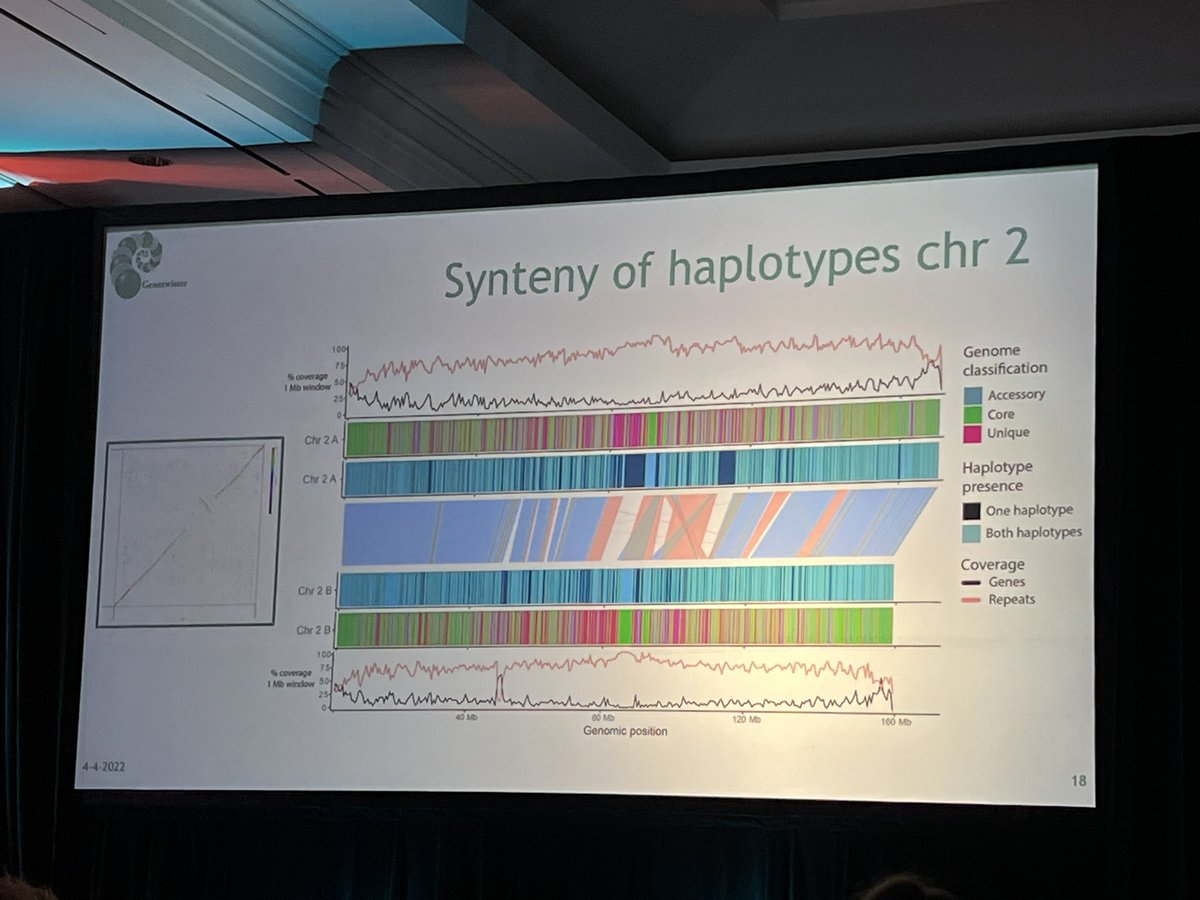
Happening now at #AGBTAG22: Bart Nijland @genetwister discusses #pangenomics (see suggested tools) and how highly contiguous, haplotype-aware genome 🧬 assemblies are more accessible than ever. @PacBio @PhaseGenomics



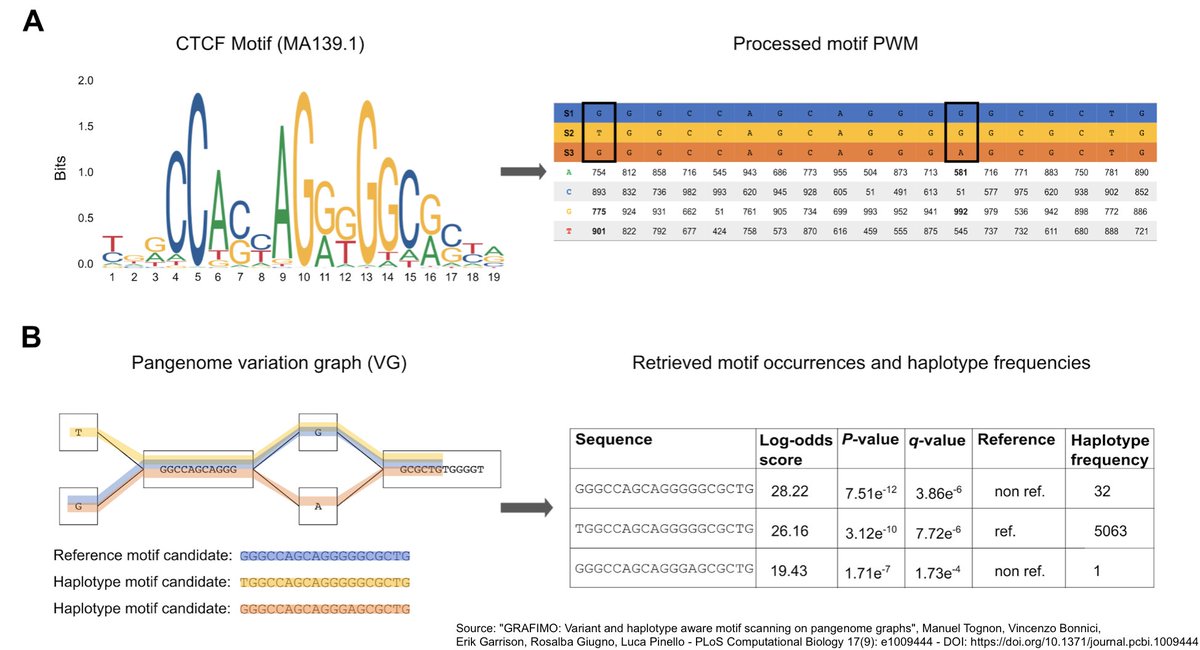
#Pangenomics #Bioinformatics: researchers have designed a computational tool for detecting transcription factor binding sites in pangenome variation graphs. The software tool could help to predict which transcription factors can bind regulatory DNA regions journals.plos.org/ploscompbiol/a…
Pangenomics and single-cell transcriptomics uncover the genetic basis of continuous bearing trait in grapevine. #Pangenomics #GrapevineGenome #ContinuousBearing @Hortres academic.oup.com/hr/advance-art…

Hot times at our 3-day #RomePanG24 course in #Rome! 🌍Participants are deeply engaged in hands-on activities, learning to leverage #pangenomics! 🧬💻 #Pangenome #Assembly #PangenomeGraphs @erikgarrison @flavia_villani




"Critical assessment of #pangenomics of metagenome-assembled genomes" by Tang Li and Yanbin Yin (@yanbinyin) highlights the importance of (and difficulties associated with) proper identification of core genes in pangenomes when working with MAGs: biorxiv.org/content/10.110…

🧬 Do you think pangenomics is hot? Attend our Workshop, Conference & Biohackathon (May 18-22, 2024). Discover genomic diversity, engage with leading scientists, and contribute to cutting-edge software. Register now! pangenome.github.io/MemPanG24/ #Pangenomics #Bioinformatics #MemPanG24

PhD candidate @dirkjan_vw studies Lactuca pangenomics in @LettuceKnow. He enjoys the collaborative spirit of the consortium where everyone supports each other. While he does spend his working days behind the computer, he also takes care of his own kitchen garden. #PanGenomics

Pandora: #nucleotide-resolution #bacterial #pangenomics with reference #graphs doi.org/10.1186/s13059… > #Genomics #Bioinformatics #Bioinformática
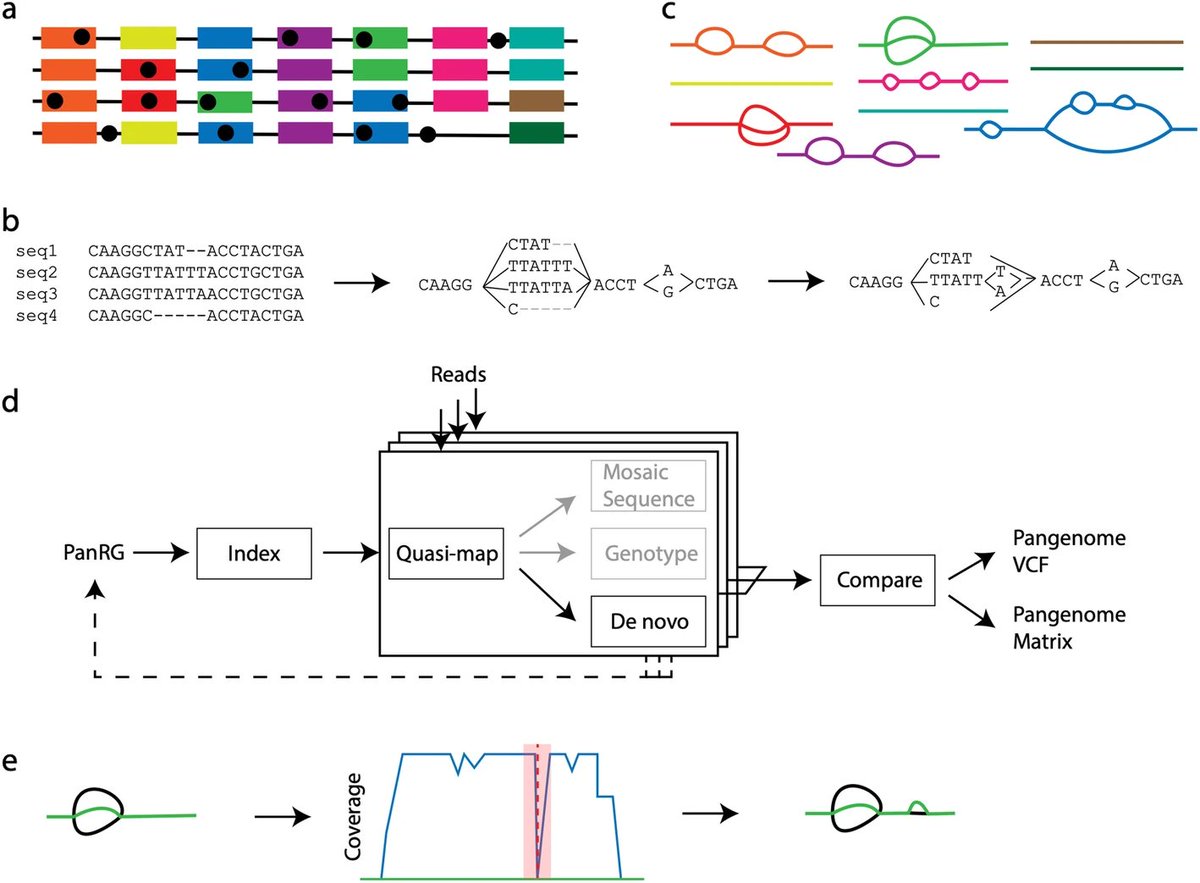
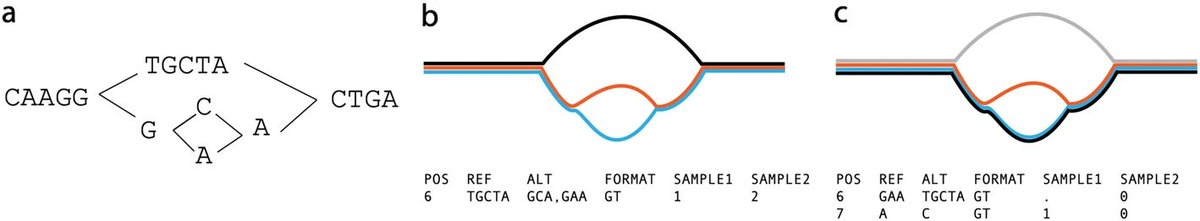
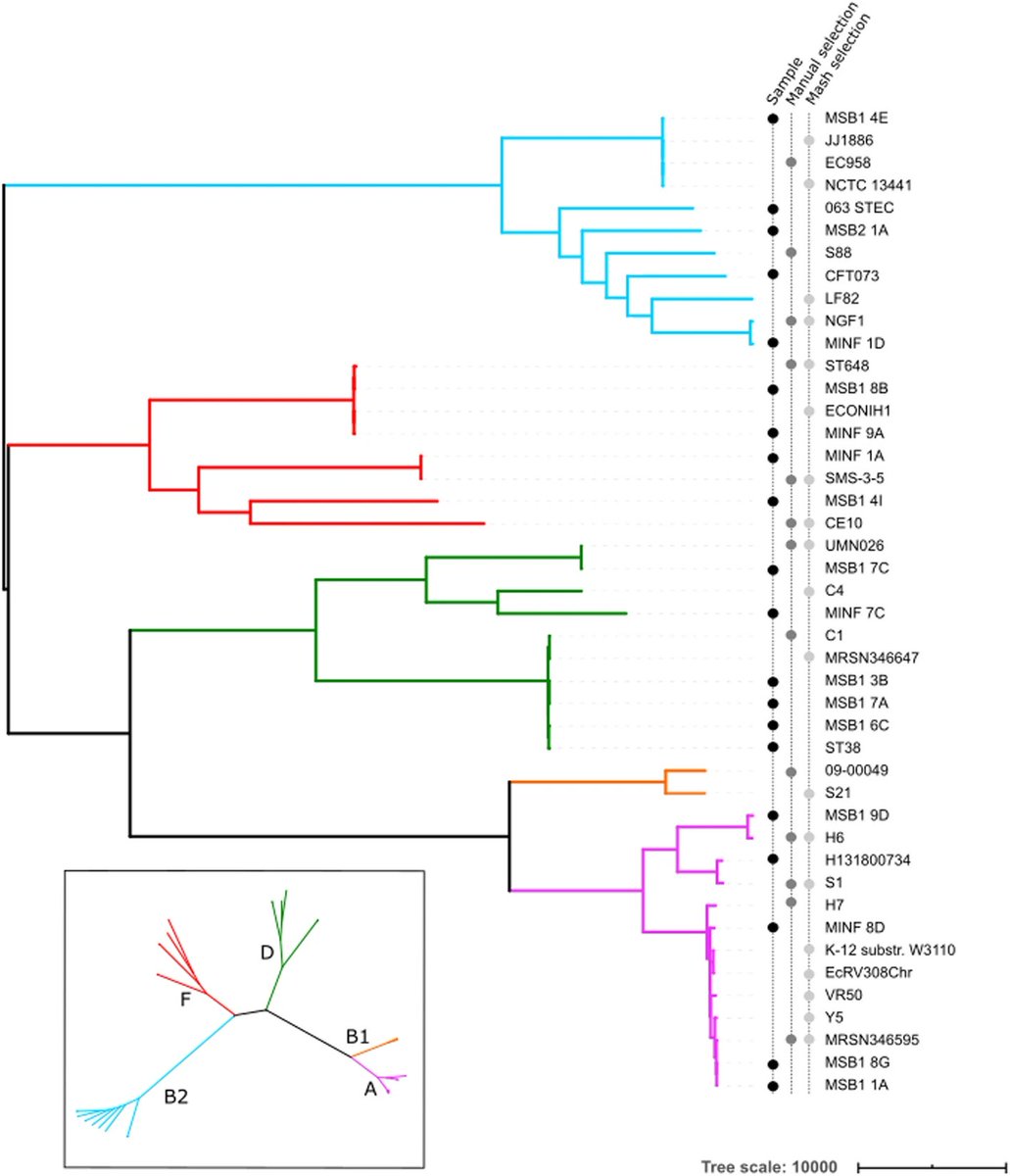
Memphis Pangenomics days, 2024 edition! Pangenome Workshop (May 18-19), Conference (May 20), & Biohackathon (May 21-22). pangenome.github.io/MemPanG24/ #Pangenomics #Bioinformatics #MemPanG24

Beginning of #ISMBECCB2023. I'm looking forward to presenting PANORAMA on Wednesday at the Function session and with my poster. @GenoLabgem @GenoUMR_GM #pangenomics

Reference-based QUantification Of gene-Dispensability (QUOD) is now available on #bioRxiv doi.org/10.1101/2020.0… @biorxiv_bioinfo @GradSchool_DILS @CeBiTec #pangenomics #Python #Bioinformatics #phdlife #homeoffice

Billi: Provably Accurate and Scalable Bubble Detection in Pangenome Graphs doi.org/10.1101/2025.1… Pangenome of cultivated beet and crop wild relatives reveals parental relationships of a tetraploid wild beet doi.org/10.1101/2023.0… #Genomics #Bioinformatics #Pangenomics
GrAnnoT, a tool for efficient and reliable annotation transfer through pangenome graph doi.org/10.1101/2025.0… KIPEs3: Automatic annotation of biosynthesis pathways doi.org/10.1101/2022.0… #FunctionalGenomics #PlantSci #Pangenomics
This meeting report from the 2024 EMBO Lecture Course on Evolutionary and Comparative Genomics highlights emerging trends across #genomeevolution, #AIapplications, #pangenomics, transposon dynamics, and the genetic basis of adaptation. The paper synthesizes discussions from…
A Pangenomic Method for Establishing a Somatic Variant Detection Resource in HapMap Mixtures.#SomaticVariantDetection #HapMap #Pangenomics @biorxivpreprint biorxiv.org/content/10.110…
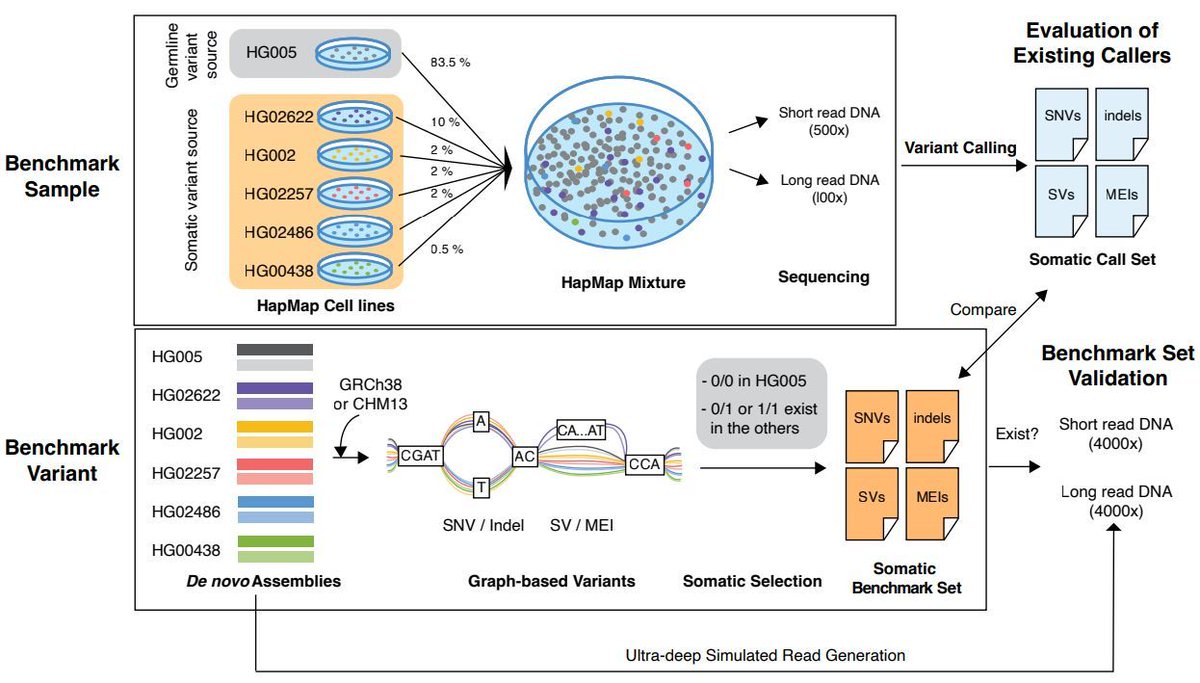
9/ Finally, thank you to everyone at @SMaHTnetwork and @NIH for supporting this incredible project. We’re excited to see how this benchmarking resource advances the field of #pangenomics and #somatic #mosaicism! 🧬💪
8/ Huge kudos to lead authors @Nahyun_Kong, @Chloe12079819, and Andrew Ruttenberg! 🎉Honored to co-supervise this work with the amazing @twang5 @WashUGenetics @GenomeInstitute. Learned so much from the brilliant minds in the @SMaHTnetwork. 💡🙌🏥🧬
biorxiv.org/content/10.110… After spotting weird gene‐clustering and 'pangenome' quirks, I dug deep and found it’s not just one tool. It’s how we misuse and misassume how 'black-box' clustering and pangenome tools work. #pangenomics #bioinformatics #geneclustering
Compressing and accelerating exabytes of pangenomic data - Summer School & Hackathon docs.google.com/presentation/d… #pangenomics

Don’t miss the first edition of the SIBE Summer School in Ferrara from 7th to 11th September 2025! Registrations are open until 20th of April! #genomics #pangenomics #conservation @sibe_iseb

Want to level up in #pangenomics? Join our workshop, conference & biohackathon in #Memphis, May 11-15, 2025. Connect with leading scientists, embrace genomic diversity, and contribute to cutting-edge software. Register now! pangenome.github.io/MemPanG25/ #Bioinformatics #MemPanG25

Improved pangenomic classification accuracy with chain statistics. #Pangenomics @biorxivpreprint biorxiv.org/content/10.110…
If you're not yet "building #pangenome graphs" to analyze your #pangenomes, now is the time to start and unlock #pangenomics true power!
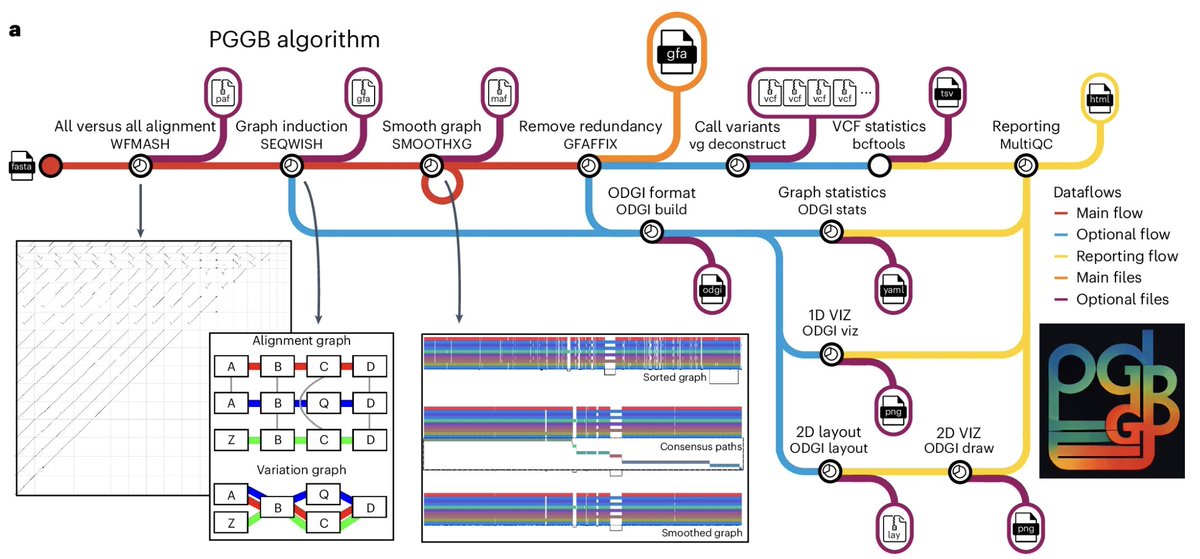
PanGenome Graph Builder is a modular framework for efficiently building unbiased pangenome graphs, supporting diverse downstream analyses. @erikgarrison @AndresGuarahino nature.com/articles/s4159…
📊 Interested in learning more about #pangenomics and topological data analysis? The open-source materials for the genomic analysis workshop are available here: carpentries-incubator.github.io/pangenomics-wo… #OpenScience #Bioinformatics #Education
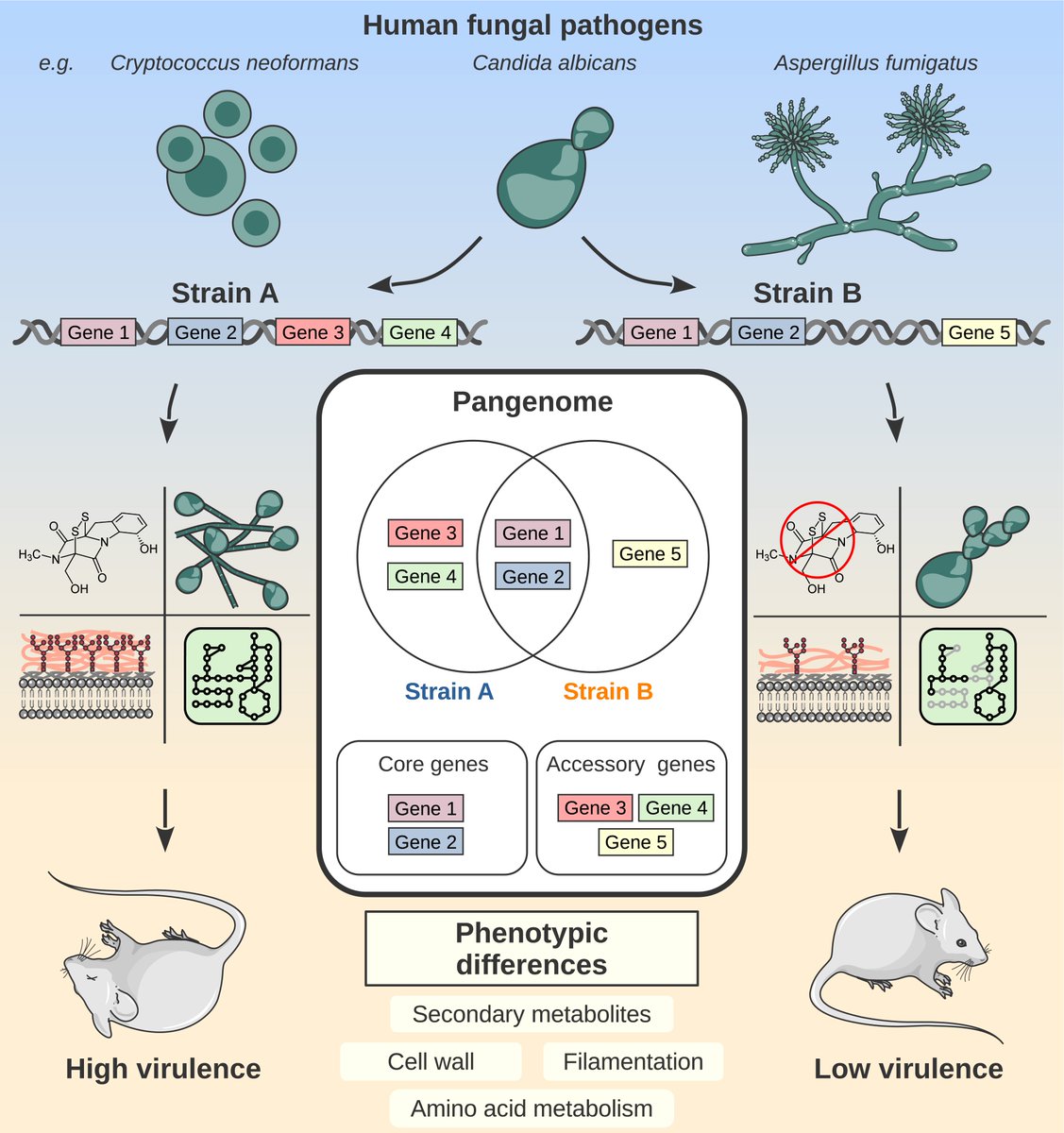
"Unraveling the #genomic diversity and virulence of human #fungal pathogens through #pangenomics" from the @AmeliaBarberPhD lab #fungi #science #bio
Personalized pangenome references. #Pangenomics #PersonalizedPangenomes #PangenomeGraphs #VariantGenotyping @naturemethods nature.com/articles/s4159…
Meeting the Challenge of Genomic Analysis: A Collaboratively Developed Workshop for Pangenomics and Topological Data Analysis. #Pangenomics #GenomeAnalysis #Training #ComputationalBiology @BioinfoAdv academic.oup.com/bioinformatics…
academic.oup.com
Meeting the challenge of genomic analysis: a collaboratively developed workshop for pangenomics and...
AbstractMotivation. As genomics data analysis becomes increasingly intricate, researchers face the challenge of mastering various software tools. The rise
Alexandra CALTEAU is presenting LABGeM developments on #pangenomics @MIGGS_workshop in Lille
We're offering two master internships to work on #pangenomics and #archaea #phylogenomics labgem.genoscope.cns.fr/category/jobs/ Come and work with us!
labgem.genoscope.cns.fr
Job Offers Archives - LABGeM
Job Offers Archives - LABGeM
🧬 Discover how Aleix Canalda Baltrons is uncovering structural variations in Mycobacterium tuberculosis through pangenomics! Learn about the novel graph approach enhancing SV detection and its implications for TB biology. #MicroSeq2024 #Microbiology #Pangenomics @TheDohertyInst

🏥Tune in to #MicroSeq2024 to hear from Alexandra-Maria Blejusca. She reveals a genomic approach for identifying different Burkholderia spp. in public health laboratories! Burkholderia diversity in Victoria & globally was also characterised using #phylogenetics and #pangenomics.

Something went wrong.
Something went wrong.
United States Trends
- 1. #GMMTV2026 992K posts
- 2. MILKLOVE BORN TO SHINE 186K posts
- 3. Good Tuesday 22K posts
- 4. WILLIAMEST MAGIC VIBES 22.2K posts
- 5. TOP CALL 9,322 posts
- 6. #WWERaw 78.1K posts
- 7. AI Alert 8,123 posts
- 8. Moe Odum N/A
- 9. Barcelona 146K posts
- 10. Brock 42.1K posts
- 11. Purdy 28.5K posts
- 12. Alan Dershowitz 2,818 posts
- 13. Check Analyze 2,421 posts
- 14. Bryce 21.3K posts
- 15. Token Signal 8,599 posts
- 16. Keegan Murray 1,558 posts
- 17. Dialyn 8,012 posts
- 18. Timberwolves 3,935 posts
- 19. Market Focus 4,644 posts
- 20. Finch 14.9K posts