#spatialdlpfc نتائج البحث
A long day of travel ✈️🚉🚕 but got myself and my poster all the way to the UK for #GBD23! 🇬🇧 Excited to present at the first poster session on #spatialDLPFC !

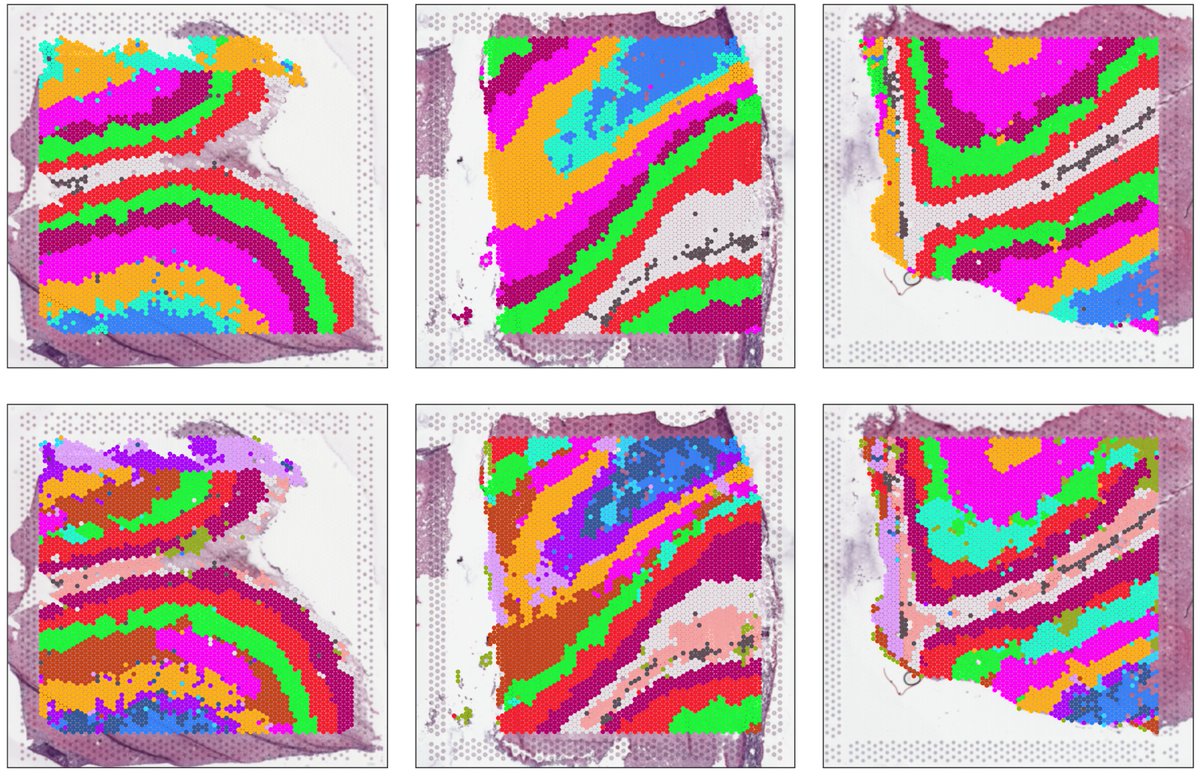
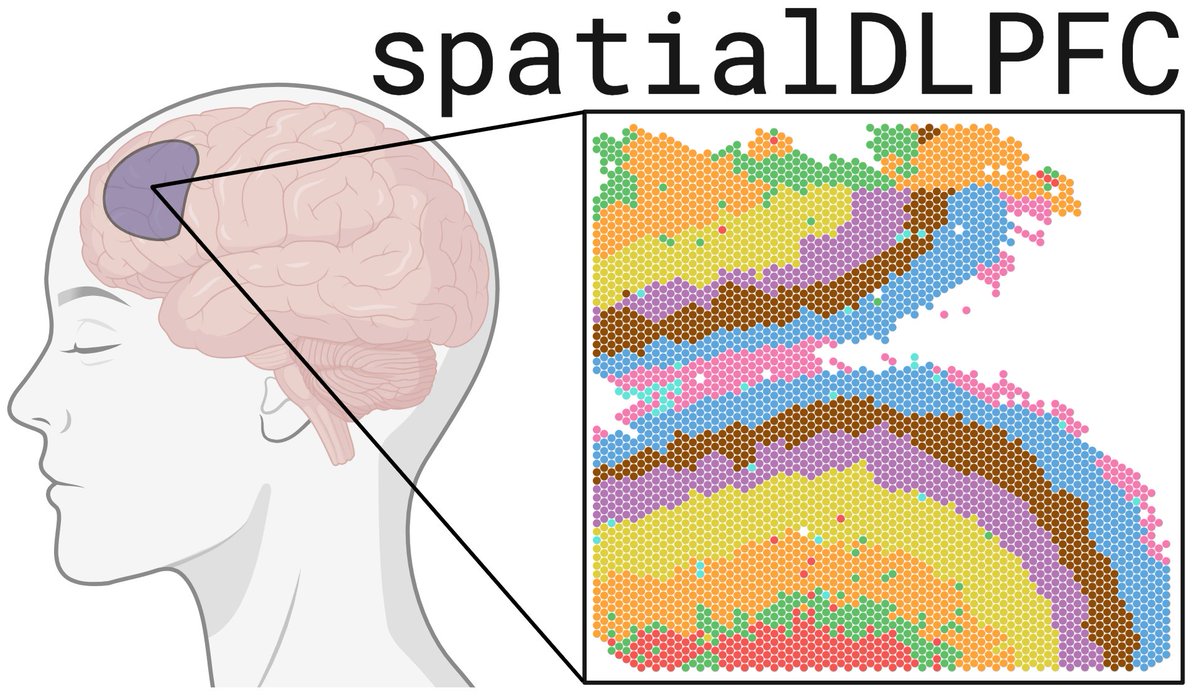
#spatialDLPFC increases ⏫ the #Visium sample size, and adds paired #snRNAseq 🎨🧪 We leverage data-driven clustering from #BayesSpace to define novel spatial domains. Also added #VisiumSPG to spatially map cell types 🗺️

Wow very proud to have won the poster award at #Connectome2024 ! 🏆 Thanks to everyone who stopped by to hear about spatial transcriptomics in the human brain 🧠 #spatialDLPFC

Poster prize, rewarding excellent poster design, explanation and discussion of results or methodology 🥇 @lahuuki for “A data-driven single cell and spatial transcriptomic map of the human prefrontal cortex”
Hot of the pre-print press! 🔥 Our latest work #spatialDLPFC pairs #snRNAseq and #Visium spatial transcriptomic data in the human #DLPFC building a neuroanatomical atlas of this critical brain region 🧠 @LieberInstitute @10xGenomics #scitwitter 📰 doi.org/10.1101/2023.0…
New in @sciencemagazine: our work from @LieberInstitute #spatialDLPFC applies #snRNAseq and #Visium spatial transcriptomic in the DLPFC to better understand anatomical structure and cellular populations in the human brain #PsychENCODE doi.org/10.1126/scienc…
Hot of the pre-print press! 🔥 Our latest work #spatialDLPFC pairs #snRNAseq and #Visium spatial transcriptomic data in the human #DLPFC building a neuroanatomical atlas of this critical brain region 🧠 @LieberInstitute @10xGenomics #scitwitter 📰 doi.org/10.1101/2023.0…

#BioC2023 Be sure to check out Nick Eagles' (github.com/Nick-Eagles) from @LieberInstitute talk on spot deconvolution benchmark + implementation in our #spatialDLPFC data at 10am 🔍

¡Gracias Pilar @pilaralvjer & Cornelis @cornelisblauw for the invitation to present our work today at the 2023 NIA/LNG Neurodegenerative Diseases Seminar Series! 🛝 by @lahuuki: speakerdeck.com/lahuuki/2023-q… mine: speakerdeck.com/lcolladotor/20… #spatialDLPFC #Visium_SPG_AD @LieberInstitute

Expect some #spatialDLPFC tweetorials soon ;) @biorxivpreprint #rstats @Bioconductor @10xGenomics

Integrated single cell and unsupervised spatial transcriptomic analysis defines molecular anatomy of the human dorsolateral prefrontal cortex biorxiv.org/cgi/content/sh… #bioRxiv
I had so much fun generating the data, and of-course being part of this incredible team and #spatialDLPFC project. To read more about spot deconvolution, check out👇
Downstream analysis were powered up 🦸 with the paired datasets 🤝. Nick Eagles (github.com/Nick-Eagles) performed a spot deconvolution benchmark analysis between #tangram #Cell2location and #SPOTlight with the #VisiumSPG data

🚀 Motivating problem: In our #spatialDLPFC project, we identified colocalized genes and wanted to map them to spatial domains (cortex layers). 🤨Is it possible to visualize the two variables in a unified spatial map instead of two side-by-side plots?

Also leveraging the paired #snRNAseq and #Visium data @BoyiGuo & @mgrantpeters studied schizophrenia associated ligand-receptor pairs with spatial context in the DLPFC 🧠

A huge thanks to all of the collaborators on the #spatialDLPFC project! Your expertise from the wet bench to the computational work made this work possible from @LieberInstitute, @jhubiostat, @ucl, and #PsychENCODE @NIMHgov 👨🔬🎉👩🔬
Looking for something fun on #FluorescenceFriday? Check out our #VisiumSPG data on samuibrowser.com to explore our spatially resolved human DLPFC transcriptomes! #spatialDLPFC @LieberInstitute

Hot of the pre-print press! 🔥 Our latest work #spatialDLPFC pairs #snRNAseq and #Visium spatial transcriptomic data in the human #DLPFC building a neuroanatomical atlas of this critical brain region 🧠 @LieberInstitute @10xGenomics #scitwitter 📰 doi.org/10.1101/2023.0…

I'm very excited that I was able to contribute to this amazing #spatialDLPFC study! We used #VistoSeg for generating cell counts which was used heavily for downstream analysis and spot deconvolution. doi.org/10.1101/2021.0…
Hot of the pre-print press! 🔥 Our latest work #spatialDLPFC pairs #snRNAseq and #Visium spatial transcriptomic data in the human #DLPFC building a neuroanatomical atlas of this critical brain region 🧠 @LieberInstitute @10xGenomics #scitwitter 📰 doi.org/10.1101/2023.0…

Thanks for the chance to present about #spatialDLPFC 🧠😁! Checkout our pre-print on #snRNAseq & #visium data in the human brain: biorxiv.org/content/10.110…
Who would have thought there could be more *layers* of complexity to cortical annotation than what was first described by Santiago Ramon y Cajal in circa 1911? Very exciting work! #spatialDLPFC biorxiv.org/content/10.110…
Congrats Nick for successfully completing your 1st🥇 @LieberInstitute seminar 🙌🏽 Nick presented #spatialDLPFC spot deconvolution results @10xGenomics #snRNAseq #Visium #VisiumSPG 🛝 speakerdeck.com/nickeagles/lib… Interact with the data research.libd.org/spatialDLPFC/#… youtu.be/cCUHRdI4NSc

Slides for part 1 on #spatialDLPFC here 🧠 speakerdeck.com/lahuuki/2023-q…
Excited to share our latest spatial transcriptomics adventure generating a data-driven molecular atlas of the human DLPFC. Excellent tweetorial from @lahuuki below. Congrats to the #spatialDLPFC team and all collaborators who contributed.
Hot of the pre-print press! 🔥 Our latest work #spatialDLPFC pairs #snRNAseq and #Visium spatial transcriptomic data in the human #DLPFC building a neuroanatomical atlas of this critical brain region 🧠 @LieberInstitute @10xGenomics #scitwitter 📰 doi.org/10.1101/2023.0…

🧠New pre-print from Louise A. Huuki-Myers et al - contributions from Mina and Melissa @PD_Progression and @PD_BrainMap #spatialDLPFC doi.org/10.1101/2023.0…
Wow very proud to have won the poster award at #Connectome2024 ! 🏆 Thanks to everyone who stopped by to hear about spatial transcriptomics in the human brain 🧠 #spatialDLPFC

Poster prize, rewarding excellent poster design, explanation and discussion of results or methodology 🥇 @lahuuki for “A data-driven single cell and spatial transcriptomic map of the human prefrontal cortex”
Thanks for the opportunity to share about #spatialDLPFC ! 🧠
The original idea formed during the collab with @LieberInstitute on #spatialDLPFC project and expanded in ongoing study led by me and @sanghokwon17 from @martinowk lab for an even larger #SRT dataset studying neuropsychiatric disorders. #TeamScienceWins x.com/lahuuki/status…
New in @sciencemagazine: our work from @LieberInstitute #spatialDLPFC applies #snRNAseq and #Visium spatial transcriptomic in the DLPFC to better understand anatomical structure and cellular populations in the human brain #PsychENCODE doi.org/10.1126/scienc…

Thanks to all #spatialDLPFC coauthors from @LieberInstitute @jhubiostat & @ucl🧑🔬 @abspangler @sanghokwon17 @BoyiGuo @mgrantpeters @HeenaDivecha @MadhaviTippani @chaichontat @anniebichnguyen @mattntran @alexisjbattle @CerceoPage @stephaniehicks @martinowk @lcolladotor @kr_maynard
New in @sciencemagazine: our work from @LieberInstitute #spatialDLPFC applies #snRNAseq and #Visium spatial transcriptomic in the DLPFC to better understand anatomical structure and cellular populations in the human brain #PsychENCODE doi.org/10.1126/scienc…

Hot of the pre-print press! 🔥 Our latest work #spatialDLPFC pairs #snRNAseq and #Visium spatial transcriptomic data in the human #DLPFC building a neuroanatomical atlas of this critical brain region 🧠 @LieberInstitute @10xGenomics #scitwitter 📰 doi.org/10.1101/2023.0…

But actually come checkout my #spatialDLPFC poster this afternoon for even more #spatialLIBD content 😁 #BioC2023
#BioC2023 Be sure to check out Nick Eagles' (github.com/Nick-Eagles) from @LieberInstitute talk on spot deconvolution benchmark + implementation in our #spatialDLPFC data at 10am 🔍

Looking forward to #BioC2023 ! @lcolladotor and I will be demoing the #spatialLIBD package 💻🔧 and I will be presenting new data and findings in my poster on #spatialDLPFC 🧠🧬 from our preprint 👩🔬 🔗doi.org/10.1101/2023.0…
**Speaker Spotlight** Louise Huuki-Myers Website: buff.ly/44gTkCz Twitter: @lahuuki Her #BioC2023 talk will be on Analyzing Spatially-Resolved Transcriptomics Data from Visium using spatialLIBD Registration for BioC2023: buff.ly/3nKEq72

Congrats Nick for successfully completing your 1st🥇 @LieberInstitute seminar 🙌🏽 Nick presented #spatialDLPFC spot deconvolution results @10xGenomics #snRNAseq #Visium #VisiumSPG 🛝 speakerdeck.com/nickeagles/lib… Interact with the data research.libd.org/spatialDLPFC/#… youtu.be/cCUHRdI4NSc

¡Gracias Pilar @pilaralvjer & Cornelis @cornelisblauw for the invitation to present our work today at the 2023 NIA/LNG Neurodegenerative Diseases Seminar Series! 🛝 by @lahuuki: speakerdeck.com/lahuuki/2023-q… mine: speakerdeck.com/lcolladotor/20… #spatialDLPFC #Visium_SPG_AD @LieberInstitute

We also used #spatialDLPFC by @lahuuki @lcolladotor @LieberInstitute. Great for #spatialregistration: research.libd.org/spatialDLPFC/#…
Slides for part 1 on #spatialDLPFC here 🧠 speakerdeck.com/lahuuki/2023-q…
speakerdeck.com
2023 Queen Square Institute of Neurology: Spatial DLPFC
Paired talk with: https://speakerdeck.com/lcolladotor/2023-queen-square-institute-of-neurology
For our last seminar 🇬🇧 trip, on Thursday May 25 @lahuuki & I will do a joint presentation on "Harnessing the power of spatially-resolved transcriptomics one step at a time" at @UCLIoN 9:30 am @LieberInstitute🧠 #spatialLIBD #spatialDLPFC #Visium_SPG_AD speakerdeck.com/lcolladotor/20…

If you'll be at @TheCrick on Tuesday 5/23 & want to learn about 🧠 RNA-seq & spatial omics deconvolution with snRNA-seq, join us at 1pm! @LabGandhi @LieberInstitute 🛝 speakerdeck.com/lcolladotor/cr… #deconvochallenge -> #TREG #DeconvoBuddies #spatialLIBD -> #spatialDLPFC #Visium_SPG_AD

Thanks to #GBD23 @lahuuki & I can finally meet in person with our #spatialDLPFC collaborators @mgrantpeters @MinaRyten (not her account or is it?🤔) + celebrate our @biorxivpreprint 📜 with them ^^ Congrats Mina on your new job! 🎉 📸: @pilaralvjer thx!

Hot of the pre-print press! 🔥 Our latest work #spatialDLPFC pairs #snRNAseq and #Visium spatial transcriptomic data in the human #DLPFC building a neuroanatomical atlas of this critical brain region 🧠 @LieberInstitute @10xGenomics #scitwitter 📰 doi.org/10.1101/2023.0…

A long day of travel ✈️🚉🚕 but got myself and my poster all the way to the UK for #GBD23! 🇬🇧 Excited to present at the first poster session on #spatialDLPFC !

#Visium_SPG_AD with @sowmyapartybun benefited from simultaneous updates to #spatialLIBD that were also needed for #spatialDLPFC by @lahuuki @abspangler Nick-Eagles et al It wouldn't have been possible without #VistoSeg v2 by @MadhaviTippani + a whole village, thanks everyone!!
🚀 Motivating problem: In our #spatialDLPFC project, we identified colocalized genes and wanted to map them to spatial domains (cortex layers). 🤨Is it possible to visualize the two variables in a unified spatial map instead of two side-by-side plots?

Also leveraging the paired #snRNAseq and #Visium data @BoyiGuo & @mgrantpeters studied schizophrenia associated ligand-receptor pairs with spatial context in the DLPFC 🧠

I'm very excited that I was able to contribute to this amazing #spatialDLPFC study! We used #VistoSeg for generating cell counts which was used heavily for downstream analysis and spot deconvolution. doi.org/10.1101/2021.0…
Hot of the pre-print press! 🔥 Our latest work #spatialDLPFC pairs #snRNAseq and #Visium spatial transcriptomic data in the human #DLPFC building a neuroanatomical atlas of this critical brain region 🧠 @LieberInstitute @10xGenomics #scitwitter 📰 doi.org/10.1101/2023.0…

Full history + archive on @ZENODO_ORG the preprint version & published one See github.com/LieberInstitut… by @lahuuki @abspangler me, et al #spatialDLPFC Full history allows you to see contributions from everyone too. Including early exploratory work like what @artaseyedian did
Hot of the pre-print press! 🔥 Our latest work #spatialDLPFC pairs #snRNAseq and #Visium spatial transcriptomic data in the human #DLPFC building a neuroanatomical atlas of this critical brain region 🧠 @LieberInstitute @10xGenomics #scitwitter 📰 doi.org/10.1101/2023.0…

A long day of travel ✈️🚉🚕 but got myself and my poster all the way to the UK for #GBD23! 🇬🇧 Excited to present at the first poster session on #spatialDLPFC !

¡Gracias Pilar @pilaralvjer & Cornelis @cornelisblauw for the invitation to present our work today at the 2023 NIA/LNG Neurodegenerative Diseases Seminar Series! 🛝 by @lahuuki: speakerdeck.com/lahuuki/2023-q… mine: speakerdeck.com/lcolladotor/20… #spatialDLPFC #Visium_SPG_AD @LieberInstitute

New in @sciencemagazine: our work from @LieberInstitute #spatialDLPFC applies #snRNAseq and #Visium spatial transcriptomic in the DLPFC to better understand anatomical structure and cellular populations in the human brain #PsychENCODE doi.org/10.1126/scienc…

Hot of the pre-print press! 🔥 Our latest work #spatialDLPFC pairs #snRNAseq and #Visium spatial transcriptomic data in the human #DLPFC building a neuroanatomical atlas of this critical brain region 🧠 @LieberInstitute @10xGenomics #scitwitter 📰 doi.org/10.1101/2023.0…

Expect some #spatialDLPFC tweetorials soon ;) @biorxivpreprint #rstats @Bioconductor @10xGenomics

Integrated single cell and unsupervised spatial transcriptomic analysis defines molecular anatomy of the human dorsolateral prefrontal cortex biorxiv.org/cgi/content/sh… #bioRxiv
#BioC2023 Be sure to check out Nick Eagles' (github.com/Nick-Eagles) from @LieberInstitute talk on spot deconvolution benchmark + implementation in our #spatialDLPFC data at 10am 🔍

#spatialDLPFC increases ⏫ the #Visium sample size, and adds paired #snRNAseq 🎨🧪 We leverage data-driven clustering from #BayesSpace to define novel spatial domains. Also added #VisiumSPG to spatially map cell types 🗺️

Wow very proud to have won the poster award at #Connectome2024 ! 🏆 Thanks to everyone who stopped by to hear about spatial transcriptomics in the human brain 🧠 #spatialDLPFC

Poster prize, rewarding excellent poster design, explanation and discussion of results or methodology 🥇 @lahuuki for “A data-driven single cell and spatial transcriptomic map of the human prefrontal cortex”
Thanks to #GBD23 @lahuuki & I can finally meet in person with our #spatialDLPFC collaborators @mgrantpeters @MinaRyten (not her account or is it?🤔) + celebrate our @biorxivpreprint 📜 with them ^^ Congrats Mina on your new job! 🎉 📸: @pilaralvjer thx!

Hot of the pre-print press! 🔥 Our latest work #spatialDLPFC pairs #snRNAseq and #Visium spatial transcriptomic data in the human #DLPFC building a neuroanatomical atlas of this critical brain region 🧠 @LieberInstitute @10xGenomics #scitwitter 📰 doi.org/10.1101/2023.0…

Congrats Nick for successfully completing your 1st🥇 @LieberInstitute seminar 🙌🏽 Nick presented #spatialDLPFC spot deconvolution results @10xGenomics #snRNAseq #Visium #VisiumSPG 🛝 speakerdeck.com/nickeagles/lib… Interact with the data research.libd.org/spatialDLPFC/#… youtu.be/cCUHRdI4NSc

For our last seminar 🇬🇧 trip, on Thursday May 25 @lahuuki & I will do a joint presentation on "Harnessing the power of spatially-resolved transcriptomics one step at a time" at @UCLIoN 9:30 am @LieberInstitute🧠 #spatialLIBD #spatialDLPFC #Visium_SPG_AD speakerdeck.com/lcolladotor/20…

If you'll be at @TheCrick on Tuesday 5/23 & want to learn about 🧠 RNA-seq & spatial omics deconvolution with snRNA-seq, join us at 1pm! @LabGandhi @LieberInstitute 🛝 speakerdeck.com/lcolladotor/cr… #deconvochallenge -> #TREG #DeconvoBuddies #spatialLIBD -> #spatialDLPFC #Visium_SPG_AD

Looking for something fun on #FluorescenceFriday? Check out our #VisiumSPG data on samuibrowser.com to explore our spatially resolved human DLPFC transcriptomes! #spatialDLPFC @LieberInstitute

Hot of the pre-print press! 🔥 Our latest work #spatialDLPFC pairs #snRNAseq and #Visium spatial transcriptomic data in the human #DLPFC building a neuroanatomical atlas of this critical brain region 🧠 @LieberInstitute @10xGenomics #scitwitter 📰 doi.org/10.1101/2023.0…

I had so much fun generating the data, and of-course being part of this incredible team and #spatialDLPFC project. To read more about spot deconvolution, check out👇
Downstream analysis were powered up 🦸 with the paired datasets 🤝. Nick Eagles (github.com/Nick-Eagles) performed a spot deconvolution benchmark analysis between #tangram #Cell2location and #SPOTlight with the #VisiumSPG data

🚀 Motivating problem: In our #spatialDLPFC project, we identified colocalized genes and wanted to map them to spatial domains (cortex layers). 🤨Is it possible to visualize the two variables in a unified spatial map instead of two side-by-side plots?

Also leveraging the paired #snRNAseq and #Visium data @BoyiGuo & @mgrantpeters studied schizophrenia associated ligand-receptor pairs with spatial context in the DLPFC 🧠

Something went wrong.
Something went wrong.
United States Trends
- 1. Browns 35.6K posts
- 2. Sanders 30.5K posts
- 3. Ravens 35.2K posts
- 4. Chiefs 48.5K posts
- 5. Sam Darnold 9,602 posts
- 6. Rams 23.1K posts
- 7. Mahomes 14.2K posts
- 8. Lamar 19K posts
- 9. Broncos 26.5K posts
- 10. Seahawks 23K posts
- 11. Riley Moss 3,318 posts
- 12. Gabriel 61.6K posts
- 13. Myles Garrett 5,406 posts
- 14. Mark Andrews 3,706 posts
- 15. Bo Nix 3,722 posts
- 16. Tony Romo 2,315 posts
- 17. #DawgPound 3,705 posts
- 18. Falcons 33.8K posts
- 19. Kelce 6,289 posts
- 20. Chase 106K posts

















