BABS Bioinformatics @TheCrick
@BABSBioinformat
News and comment from the core bioinformatics & biostatistics team analysing data for scientists at The Francis Crick Institute
You might like
We are hiring! We have a fantastic opportunity for a motivated bioinformatician to work at @TheCrick on a large multi-omic single-cell project in immuno-oncology. please RT 🙂 @BABSBioinformat #multiomics #singlecell #immunoOncology nature.com/naturecareers/…
We are hiring. We are looking for a senior bioinformatics analyst to join us to work on amazing data rich projects in collaboration with @TheCrick scientists. Apply today #Bioinformatics #sciencejobs crick.ac.uk/careers-study/…
Fascinating work on spatial bias in metastatic subclone selection by @kevlitchfield, @TurajlicLab, @SwantonLab and Paul Bates's group. Stuart Horswell providing the bioinformatics. Nice to see our long-term collaboration with these groups continues to be fruitful. Congrats all.
📰 A multidisciplinary team of researchers led by @kevlitchfield at @uclcancer and @TurajlicLab, @SwantonLab and Paul Bates at the Crick has found that the cancer cells in the centre of tumours have the highest chance of spreading around the body. crick.ac.uk/news/2021-05-1…
More great @nf_core work here from our own @aka_hpatel. This time a new release of nf-core/rnaseq v3.1. The pipelines just keep coming. Congrats to Harshil, all involved and the wider nf-core community for crafting these incredibly useful pipelines.
Pipeline release! nf-core/rnaseq v3.1 (RNA sequencing analysis pipeline using STAR, RSEM, HISAT2 or Salmon with gene/isoform counts and extensive quality control.) See the changelog: github.com/nf-core/rnaseq…
I would like to present our updated pipeline for de novo assembly and intra-host/low-frequency variant calling for viral samples from metagenomics or amplicon-based preps. This has been quite a while coming now but hopefully it was worth the wait! A 🧵👇

Pipeline release! nf-core/viralrecon v2.0 (Assembly and intrahost/low-frequency variant calling for viral samples) See the changelog: github.com/nf-core/viralr…
Important work here showing the effectiveness of #SARSCoV2 PCR testing in asymptomatic patients over time from @AdamJKucharski and others with statistical input from our own @gavinpaulkelly. Congratulations to all involved #CovidTesting
How does PCR detectability vary over time since infection, and what are the implications for routine testing? Analysis with @HellewellJoel @timwrussell @bealelab @SAFERuclh and others now peer-reviewed and published: bmcmedicine.biomedcentral.com/articles/10.11…



Great human spinal cord development work here with comparisons to mouse from @t_rayon and the @briscoejames lab. #scrnaseq analysis and neat visualisation by our own Chris Barrington #Bioinformatics
New #preprint and open resource on the developing human spinal cord: conservation and contrast between mouse and human @t_rayon @briscoejames Want to check your predictions directly? Checkout Chris’ handy work @BABSBioinformat shiny.crick.ac.uk/scviewer/neura… biorxiv.org/content/10.110…
We are looking for 2 senior scientists to join the Sequencing Facility @TheCrick: - Single Cell Scientist to expand our capacity and develop new applications crick.wd3.myworkdayjobs.com/External/job/L… - Joint appointment with @LabGandhi to support @ASAP_Research project crick.wd3.myworkdayjobs.com/External/job/L…
Second paper of the day for @BABSBioinformat Congratulations to @KasperFugger, the West lab @TheCrick, our own @aka_hpatel for providing the #CRISPR analysis and #Bioinformatics and all else involved.
Very excited to see our paper out in @ScienceMagazine describing the sensitisation of #BRCA cancer cells to #PARPinhibitors by targeting the ‘nucleotide sanitiser’ DNPH1. Massive thanks @SantosMariana_ @GraemeHewitt2 @BoultonLab @aka_hpatel @CarellThomas science.sciencemag.org/content/372/65…
Was an absolute pleasure collaborating on the #bioinformatics for this paper with @KasperFugger. Whole genome CRISPR screens are incredibly revealing when performed well! Congrats to all involved 🥳🤩😎✂️🧬
Very excited to see our paper out in @ScienceMagazine describing the sensitisation of #BRCA cancer cells to #PARPinhibitors by targeting the ‘nucleotide sanitiser’ DNPH1. Massive thanks @SantosMariana_ @GraemeHewitt2 @BoultonLab @aka_hpatel @CarellThomas science.sciencemag.org/content/372/65…
Great work here by the @ScaffidiLab with #CRISPR screen analysis and #Bioinformatics by our own @aka_hpatel Congratulations to all involved.
From dosage compensation in flies to cancer-specific vulnerability in humans - via replication stress. Nature is full of surprises.@TheCrick @MonserratJosep @NatureCellBio @aka_hpatel @DalalLabNCI @louise_richie nature.com/articles/s4155…

Oncogenic RAS transcriptional activity predicts outcome and response to chemotherapy in lung adenocarcinoma is the latest from a collaboration with the #downwardlab and our own @philipeast and @gavinpaulkelly with a nice thread from @sdecarne #KRAS #lungcancer #bioinformatics
Excited to share our preprint on #KRAS in #lungcancer. With @phileast, we show that oncogenic RAS transcriptional activity predicts outcome and response to chemotherapy in lung adenocarcinoma whereas KRAS mutational status does not! Please share 🙂 biorxiv.org/content/10.110…
Great work here from the @tatelab on targeting MYC deregulation in cancer. Bioinformatics support provided by our own Miriam Llorian-Sopena and Probir Chakravarty. Congratulations to all involved.
A new approach to target MYC deregulation in cancer -synthetic lethality with NMT inhibition! IMP1320 eliminates high-MYC tumours in 6 days @DinisCalado @sangerinstitute @TheCrick @impchemistry @imperialcollege @biorxivpreprint : biorxiv.org/content/10.110… #oncology #drugdiscovery
Congratulations to the Behrens lab and now BABS' alumni Stuart Horswell for their latest showing proteasomal degradation of the tumour suppressor FBW7 requires branched ubiquitylation by TRIP12. #bioinformaticsbybabs nature.com/articles/s4146…
Please check out and retweet this postdoc opportunity to work in an outstanding academic environment across UCL/Crick, and in a growing & fun lab! nature.com/naturecareers/…
Look #KRAS goes mainstream. There is even a quote from our very own Obi-Wan Kenobi @TheCrick The epic battle with cancer's 'Death Star' theguardian.com/science/2021/m…
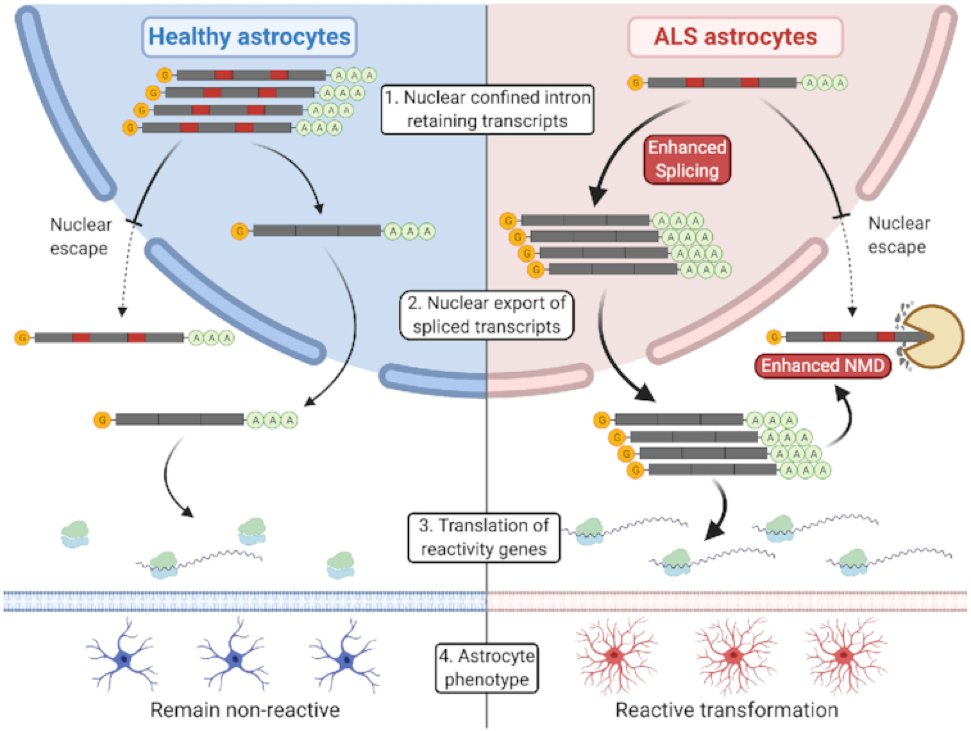
Another paper here with input from BABS and another authorship this week for our own @gavinpaulkelly Congratulations to the @PataniLab, Gavin and all involved #astrocytes #Bioinformatics
Our latest in Nucleic Acids Research (@NAR_Open) reveals how astrocytes might become harmfully reactive through a post-transcriptional mechanism: academic.oup.com/nar/advance-ar… Well done @ojziff @DoaaMTaha2 and Hamish Crerar @PataniLab @UCLIoN @TheCrick @mndresearch #DrivingMNDresearch
After a long & fruitful collab w/ the Kiyoshi Nagai lab @MRC_LMB we bring you psiCLIP - a method for profiling helicase-RNA contacts in defined spliceosome states. Led by Lisa Strittmatter & me @ule_lab @TheCrick, w/ fantastic insights from @Seb_Fica 1/7 nature.com/articles/s4146…
📰 New research in @FrontImmunol from our Tuberculosis Lab, @BABSBioinformat and @UCT_news shows why antiretroviral treatments for HIV also reduce the risk of people developing an active and dangerous Tuberculosis infection. crick.ac.uk/news/2021-03-0…
The fruits of a successful collaboration with the Tuberculosis Lab @TheCrick with our own Deborah Schneider-Luftman, Chris Barrington and @gavinpaulkelly involved. Congratulations to the Wilkinson Lab and all involved #bioinformatics #tuberculosis #HIV frontiersin.org/articles/10.33…
United States Trends
- 1. Lakers 50.7K posts
- 2. #AEWDynamite 45.8K posts
- 3. Epstein 1.5M posts
- 4. Jokic 16.3K posts
- 5. Shai 15K posts
- 6. #AEWBloodAndGuts 5,670 posts
- 7. #Survivor49 3,685 posts
- 8. Darby 5,439 posts
- 9. Thunder 42.1K posts
- 10. Kyle O'Reilly 1,863 posts
- 11. Steph 26.1K posts
- 12. Rory 7,406 posts
- 13. Kobe Sanders N/A
- 14. Moxley 2,914 posts
- 15. Spencer Knight N/A
- 16. Hobbs 28.7K posts
- 17. Blood & Guts 25.2K posts
- 18. Caruso 4,042 posts
- 19. #SistasOnBET 2,270 posts
- 20. Warriors 50K posts
Something went wrong.
Something went wrong.











![nf_core's tweet card. [3.1] - 2021-05-13 ⚠️ Major enhancements Samplesheet format has changed from group,replicate,fastq_1,fastq_2,strandedness to sample,fastq_1,fastq_2,strandedness This gives users the flexibility t...](https://pbs.twimg.com/card_img/1986647151233900544/6n1Hj68-?format=jpg&name=orig)

![nf_core's tweet card. [2.0] - 2021-05-13 ⚠️ Major enhancements Pipeline has been re-implemented in Nextflow DSL2 All software containers are now exclusively obtained from Biocontainers Updated minimum Nextflow version ...](https://pbs.twimg.com/card_img/1986422223863783424/M_aA36LF?format=jpg&name=orig)