Macas lab
@MacasLab
Interested in DNA repeats, centromeres, plant genomics and bioinformatics. Located at the @BiologyCentre of the Czech Academy of Sciences. Member of @ELIXIRCZ
You might like
Great collaboration led by Houben Lab on atypical centromeres and kinetochores in #Chionographis and #Chamaelirium plants: doi.org/10.1101/2025.0…

The program of the 12th @RepeatExplorer Workshop (May 27-29) is online: repeatexplorer.org/?page_id=1423
A new genome assembly of the pea cultivar Cameor provides resources for functional genomics and genetics: doi.org/10.1101/2025.0…
Registration open to the 12th @RepeatExplorer Workshop (May 27-29), this year focusing on repeat annotation in genome assemblies ! repeatexplorer.org/?page_id=14
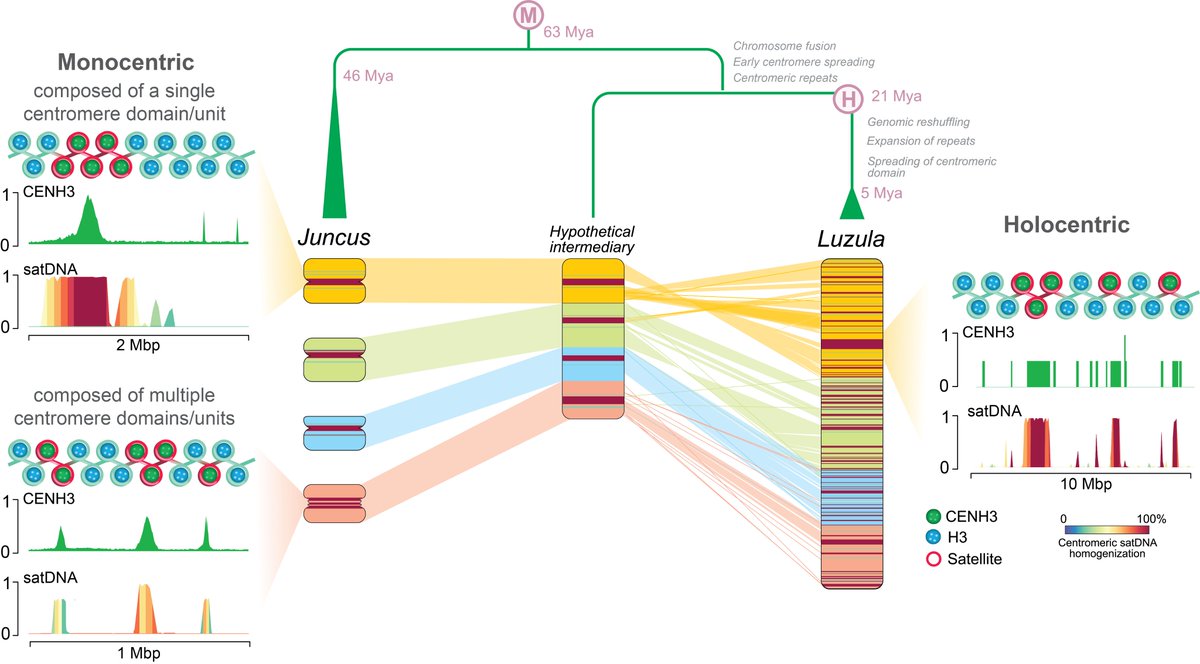
🥳Happy 🥳to share the final version now officially published in @NatureComms Repeat-based holocentromeres of the woodrush Luzula sylvatica reveal insights into the evolutionary transition to holocentricity nature.com/articles/s4146… Awesome work from @YMata_Sucre and Marie Krátká
🔥Thrilled to announce our recent work🔥 Repeat-based holocentromeres of the woodrush Luzula sylvatica reveal new insights into the evolutionary transition from mono- to holocentricity Now on bioRxiv: biorxiv.org/content/10.110… Congrats on great work from @YMata_Sucre @KratkaMarie
Excited to share our tools for LTR-retrotransposon annotation in plant genomes ! Supported by @ELIXIRCZ. doi.org/10.1093/nargab…
DANTE and DANTE_LTR: computational pipelines implementing lineage-centered annotation of LTR-retrotransposons in plant genomes. doi.org/10.1101/2024.0…
🔥Thrilled to announce our recent work🔥 Repeat-based holocentromeres of the woodrush Luzula sylvatica reveal new insights into the evolutionary transition from mono- to holocentricity Now on bioRxiv: biorxiv.org/content/10.110… Congrats on great work from @YMata_Sucre @KratkaMarie

Now published in Chromosome Res. rdcu.be/dzBlk
Targeting conserved regions of #kinetochore proteins KNL1 and NDC80 provides antibodies for labeling #centromeres across a spectrum of seed plants ! Great work by @LudCris & @PavelNeumann7 in collaboration with @YiTzuKuo, @YMata_Sucre and @marques_et_al. doi.org/10.1101/2023.1…

🚨BREAKING: Over 1,000 scientists in 14 countries hold historic demonstrations in support of gene editing. The scientists are urging MEPs to back New Genomic Techniques (#NGTs) ahead of a crunch vote in the EU parliament tomorrow. Please like and share if you believe in…
Targeting conserved regions of #kinetochore proteins KNL1 and NDC80 provides antibodies for labeling #centromeres across a spectrum of seed plants ! Great work by @LudCris & @PavelNeumann7 in collaboration with @YiTzuKuo, @YMata_Sucre and @marques_et_al. doi.org/10.1101/2023.1…

📢 Hey REPET users ! A new version of the Docker #REPET image is available! hub.docker.com/r/urgi/docker_… It includes the REXdb (@MacasLab) and Dfam (@isbsci) databases formatted for REPET use to improve the classification of your #Transposons reference library.
Great collaboration with Andreas Houben @YiTzuKuo and @MacasLab now published online in @NatureComms. Here we show that holocentromeres can also consist of only a few centromeric units with megabase-sized repeat arrays in similar ranges as in monocentromeres! Crazy!
A new type of plant holocentromere ! Excited to be part of a great team led by Houben lab at @LeibnizIPK, including @YiTzuKuo, @marques_et_al and many others. Supported by #GACR, @BiologyCentre and @ELIXIRCZ. doi.org/10.1038/s41467…
A new type of plant holocentromere ! Excited to be part of a great team led by Houben lab at @LeibnizIPK, including @YiTzuKuo, @marques_et_al and many others. Supported by #GACR, @BiologyCentre and @ELIXIRCZ. doi.org/10.1038/s41467…
Fantastic #RepeatExplorer workshop organized by @MacasLab. A huge thank you to our special guest @JohannConfais from @INRAE_URGI for his captivating presentation on REPET. Thanks to the backing of @ELIXIRCZ & @ELIXIREurope, this event marked the beginning of a new collaboration.


Our paper on aberrant kinetochores of #holocentric #Cuscuta plants just published in PNAS journal! Great work by @PavelNeumann7, @LudCris and collaborators! Supported by @ELIXIR_CZ and @BiologyCentre. doi.org/10.1073/pnas.2…
Big step forward for faba bean!! Genome sequence published in Nature. Many @ProFaba_SusCrop @suscrop partners involved. go.nature.com/3Li9kgO


Now we can map genes in #fababean! The 13Gb reference genome is published in @Nature after an amazing community effort. Thanks to all authors for great contributions! doi.org/10.1038/s41586… #legumes #plantprotein
Happy to be a part of this great team ! Supported by #GAČR, @ELIXIRCZ and @BiologyCentre
Marking a milestone in #plant #genomics!!! An amazing team of scientists around the globe unraveled the giant #fababean genome (~ 13 Gb), which is now out @Nature ..Huge congrats to all authors!!! #plantprotein @LeibnizIPK @LeibnizWGL nature.com/articles/s4158…
United States Trends
- 1. Steelers 52.1K posts
- 2. Rodgers 21.2K posts
- 3. Chargers 36.7K posts
- 4. Tomlin 8,246 posts
- 5. Schumer 221K posts
- 6. Resign 105K posts
- 7. #BoltUp 2,963 posts
- 8. #TalusLabs N/A
- 9. Tim Kaine 18.6K posts
- 10. Keenan Allen 4,858 posts
- 11. #HereWeGo 5,659 posts
- 12. #RHOP 6,884 posts
- 13. Durbin 26.2K posts
- 14. Angus King 15.7K posts
- 15. #ITWelcomeToDerry 4,506 posts
- 16. Gavin Brindley N/A
- 17. Herbert 11.7K posts
- 18. 8 Dems 6,887 posts
- 19. 8 Democrats 8,865 posts
- 20. Ladd 4,412 posts
You might like
-
 André Marques
André Marques
@marques_et_al -
 Mobile DNA
Mobile DNA
@MobDNAjournal -
 Terezie Mandakova
Terezie Mandakova
@TeriMandakova -
 Ilya Kirov
Ilya Kirov
@ilyavkirov -
 Francisco J Ruiz-Ruano
Francisco J Ruiz-Ruano
@fjruizruano -
 Diogo C. Cabral-de-Mello
Diogo C. Cabral-de-Mello
@D_CabraldeMello -
 Aleš Pečinka
Aleš Pečinka
@APecinka -
 Petr Nguyen
Petr Nguyen
@mao_626 -
 Emiliano Martí 🇦🇷
Emiliano Martí 🇦🇷
@emilianomarti7 -
 Wide hybridization group
Wide hybridization group
@Widehybridizat1 -
 Alexandros Bousios
Alexandros Bousios
@abousios
Something went wrong.
Something went wrong.