Biomni
@ProjectBiomni
A General-Purpose AI Agent to Automate Biomedical Research @Stanford
🧪Announcing @ProjectBiomni × @pylabrobot — a step toward an AI biologist that can think, design, and execute wet-lab experiments end-to-end. See Biomni in action below 👇 Super fun collab with @RetroBio_ @rickwierenga @serena2z! Learn more: biomni.stanford.edu/blog/pylabrobo…
🚀 Biomni v0.0.7 is live! New features: 📜 PDF export for agent chats 🦠 SOTA cell type transfer algorithms 🔬LazySlide pathology support 🔍Claude web search 🌟Bioimaging pipeline tools 🧬Gene conversion & ESM embeddings 🪴Glycoengineering capabilities Plus improved commercial…
this guys calenders gotta be crazy

👋Excited to announce @Sagebio ✖️ @ProjectBiomni Biomni agent can now discover using 3.74 Petabytes of Sage data to enable deeper insights into rare disease, cancer, Alzheimer's, and more 🧬Learn more: biomni.stanford.edu/blog/sage-biom…

👋Announcing @ConsensusNLP X @ProjectBiomni Literature research underpins every biomedical breakthrough. We are excited to integrate the best literature engine with access to 200M+ peer-reviewed papers to Biomni for generating deeper and more factual biomedical insights!…

🚀 Thrilled to share a preview of Biomni-R0 — we trained the first RL agent end-to-end for biomedical research. ➡️ nearly 2× stronger than its open-source base ➡️ >10% better than frontier closed-source models ➡️ Scalable path to hill climb to expert-level performance 🔗…

🚀 Biomni v0.0.6 is live! New features: 🔬 Clinicaltrials.gov integration 🧬 Copy Number Variant analysis tools added 🤖 Autonomous AI function generation 📚 Complete Sphinx documentation ⚡ Performance upgrades including centralized config management, fixed environment…
🦠Microbiome analysis usually involves complex statistical workflows to identify disease-specific taxa and diversity patterns from thousands of microbial features. Input your microbiome CSV → get novel insights into microbial biomarkers, gut-disease associations, and testable…
⚛️ Molecular docking usually involves complex software setup and structure preparation that can take hours. Input your protein target 🎯 + small molecule (SMILES) 😆 → get binding poses, affinity predictions, and interaction analysis in minutes using tools like AutoDock Vina…
Biomni open source version 0.0.5 is now released! 🎉 Since our last release, we’ve seen amazing contributions from the community: 📊 New Data Sources - OpenFDA and Monarch databases added, panhumanpy for cell type annotation, DepMap data for CRISPR screens and cancer cell lines…
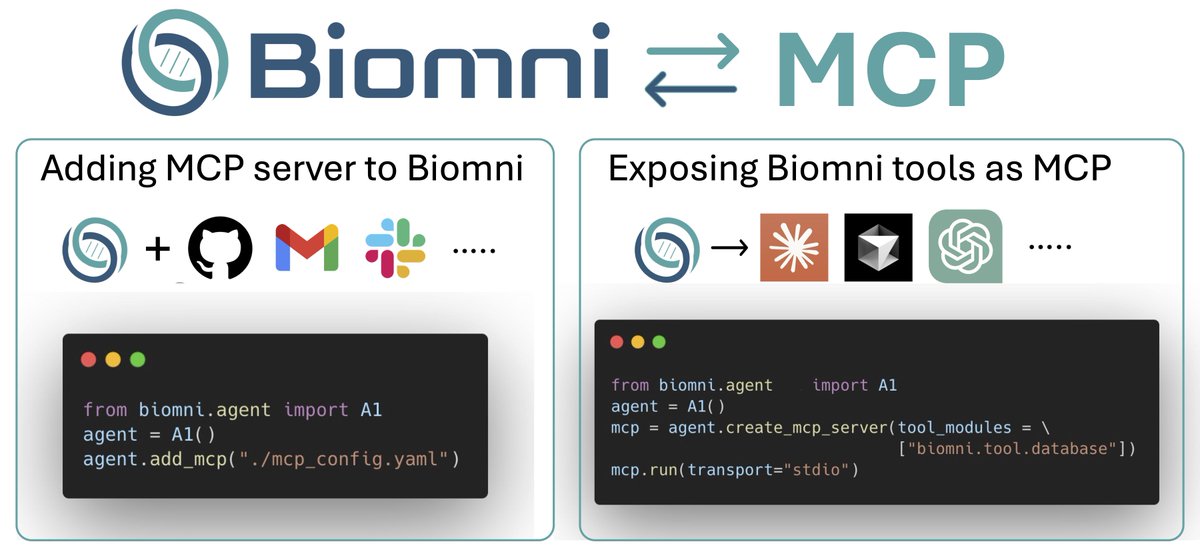
🔗@ProjectBiomni now supports MCP! Now you can have a real research assistant to share analysis results with Slack/email, upload to GitHub, etc. Biomni tools are also exposed as MCP servers! Thanks to @serena2z Shibahara Takuma Try it out: github.com/snap-stanford/…
🧪Rare diseases diagnosis requires extensive phenotype analysis, genetic variant interpretation, and literature review across multiple databases. Input patient symptoms (HPO terms) + genetic variants → get ranked differential diagnoses, pathogenicity assessments, and targeted…
We're excited to collaborate with @ProjectBiomni to bring @scverse_team tools closer to a code-free future. This is an early-stage capability, and your feedback will shape what comes next. Try it out and help us build a more accessible analysis experience.
Thrilled to share @scverse_team x @ProjectBiomni! Biomni agent now supports 10 scverse core packages including scanpy, squidpy, scirpy, pertpy, etc! You can now use natural language to unlock complex single cell, spatial, and perturbation data analysis to generate novel…
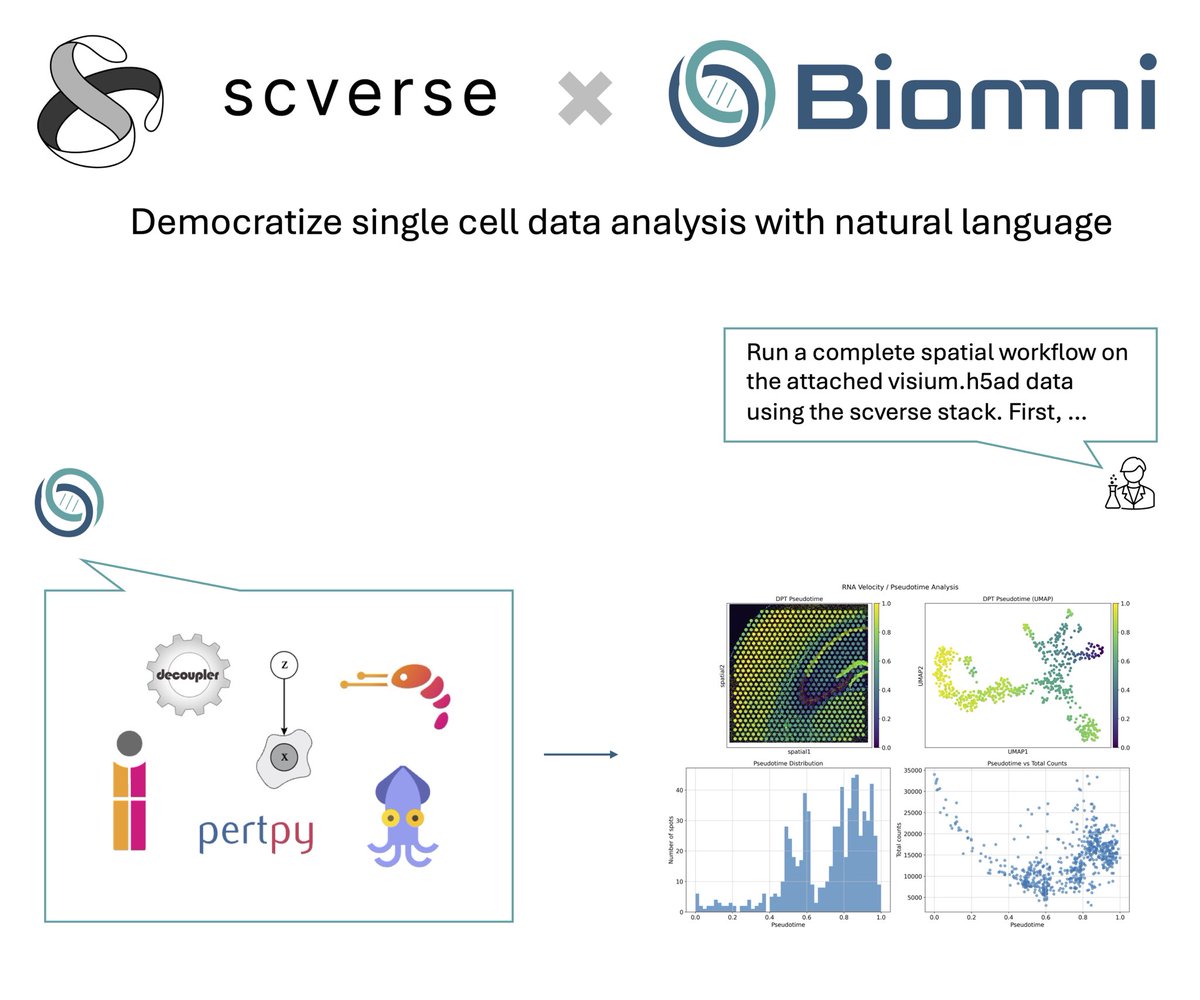
Thrilled to share @scverse_team x @ProjectBiomni! Biomni agent now supports 10 scverse core packages including scanpy, squidpy, scirpy, pertpy, etc! You can now use natural language to unlock complex single cell, spatial, and perturbation data analysis to generate novel…

🤝Excited to announce @scverse_team X @ProjectBiomni! Biomni now supports 10 core scverse packages—including scanpy, squidpy, scirpy, and pertpy—enabling researchers to perform single-cell, spatial, and perturbation analyses using natural language. biomni.stanford.edu/blog/scverse-b…

Cloning CRISPR sgRNAs into plasmids requires tedious design work—optimizing target sites, designing oligos, and creating protocols normally takes hours. Paste your target gene + backbone → get sgRNAs, oligos, and cloning protocols in minutes. 🧬
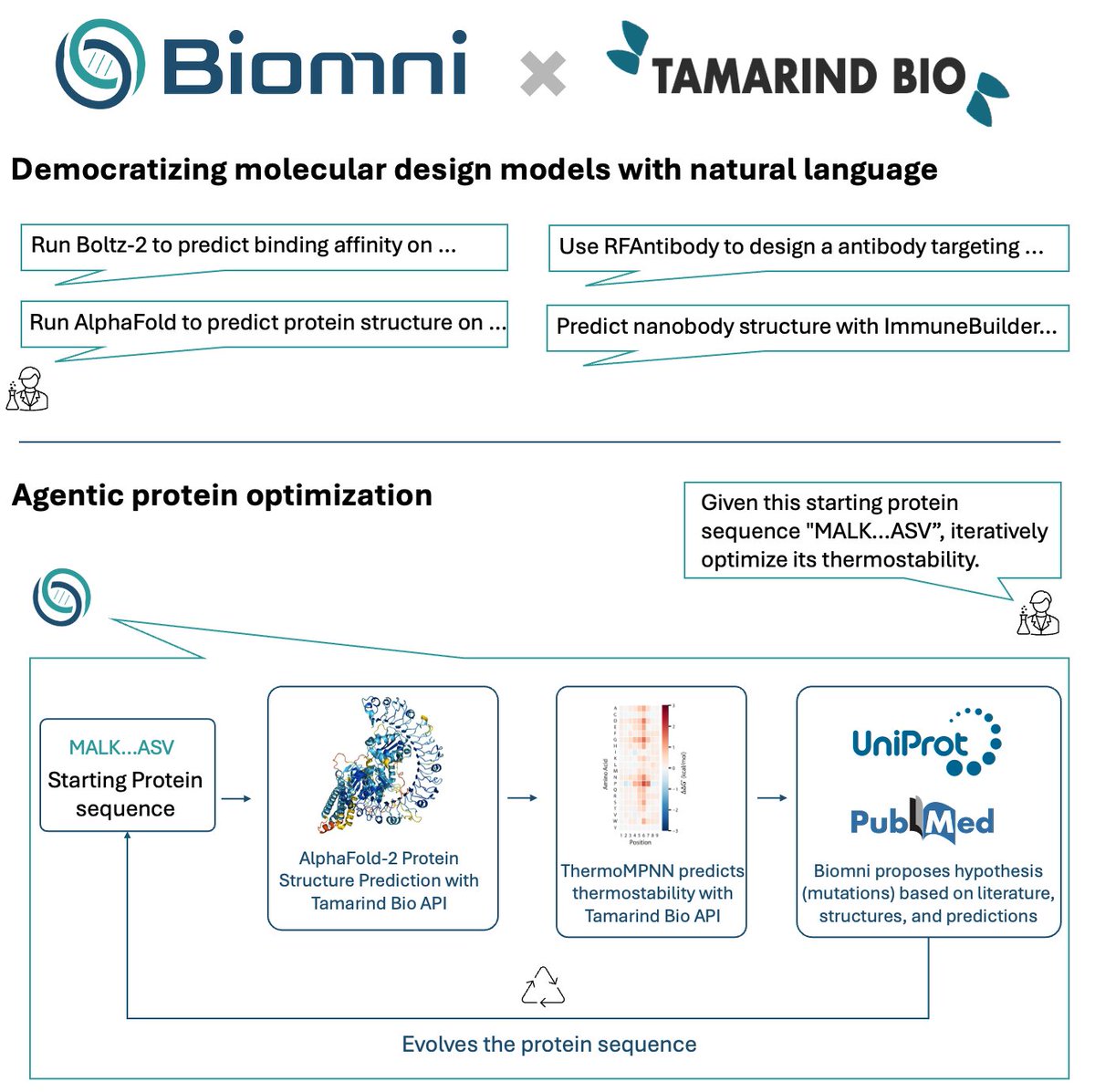
Partnering with Tamarind Bio @kavi_deniz @tamarindbio really levels up Biomni @ProjectBiomni with protein engineering capabilities. For biologists who aren't sure how to get started with computational tools - this enables all of us to access frontier protein models without any…
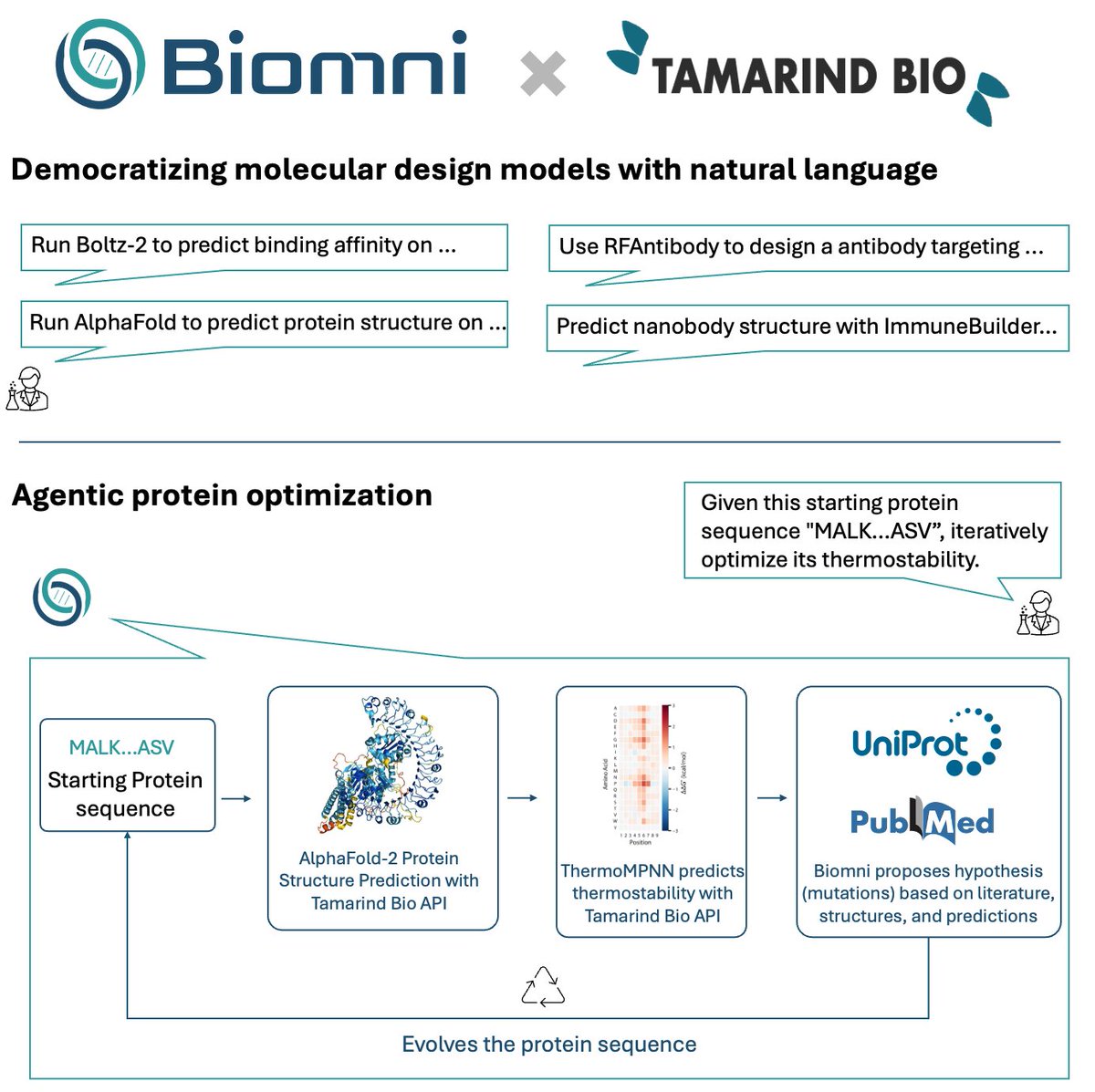
🤝Excited to partner with Tamarind @kavi_deniz to build towards an agentic AI protein designer. 🔁 Agentic protein optimization — Starting from a sequence, Biomni iteratively improves thermostability by orchestrating AlphaFold-2, ThermoMPNN, and reasoning over predictions and…
👋 Biomni v0.0.4 is now released! In one week of open-sourcing, we have seen amazing contributions from the community: - Multimodal LLM ChatNT @instadeepai as a tool to let Biomni talk with raw DNA/RNA/protein sequence - Added biomedical knowledge graph PrimeKG with millions of…
🧪Designing a CRISPR screen take days—finding gene targets, controls, and creating experimental protocols. Biomni curates target genes in minutes using advanced web search, literature mining, and pathway analysis. ⚡️ ✨Try it at biomni.stanford.edu
United States 趨勢
- 1. Cowboys 59.6K posts
- 2. Panthers 59.1K posts
- 3. Ravens 59K posts
- 4. Dolphins 41.9K posts
- 5. Browns 55.8K posts
- 6. #KeepPounding 5,574 posts
- 7. Colts 51.3K posts
- 8. Eberflus 7,288 posts
- 9. Drake Maye 17.5K posts
- 10. Steelers 58K posts
- 11. Rico Dowdle 6,482 posts
- 12. Chargers 48.5K posts
- 13. James Franklin 46.5K posts
- 14. Pickens 15.8K posts
- 15. Penn State 63.2K posts
- 16. Herbert 13.3K posts
- 17. Dillon Gabriel 3,502 posts
- 18. Saints 58.2K posts
- 19. #HereWeGo 5,411 posts
- 20. Diggs 8,798 posts
Something went wrong.
Something went wrong.