Prashant Bhat
@_prashantbhat
Uncovering the functions of genome organization | M.D. @UCLA👨🏽⚕️| Ph.D. '23 @mitchguttman @Caltech | Previously worked on #CRISPR @doudna_lab @UCBerkeley '14
You might like
A long sought after hypothesis has been that organizing biomolecules in and around nuclear bodies is essential for functional RNA processing. We now provide direct evidence of this in @Nature for nuclear speckles and mRNA splicing. Check out this thread! nature.com/articles/s4158…
What are the functional roles of 3D genome organization? Our paper in @Nature led by @_prashantbhat demonstrates how dynamic 3D genome organization around nuclear speckles plays a crucial role in regulating mRNA splicing efficiency. rdcu.be/dHkez
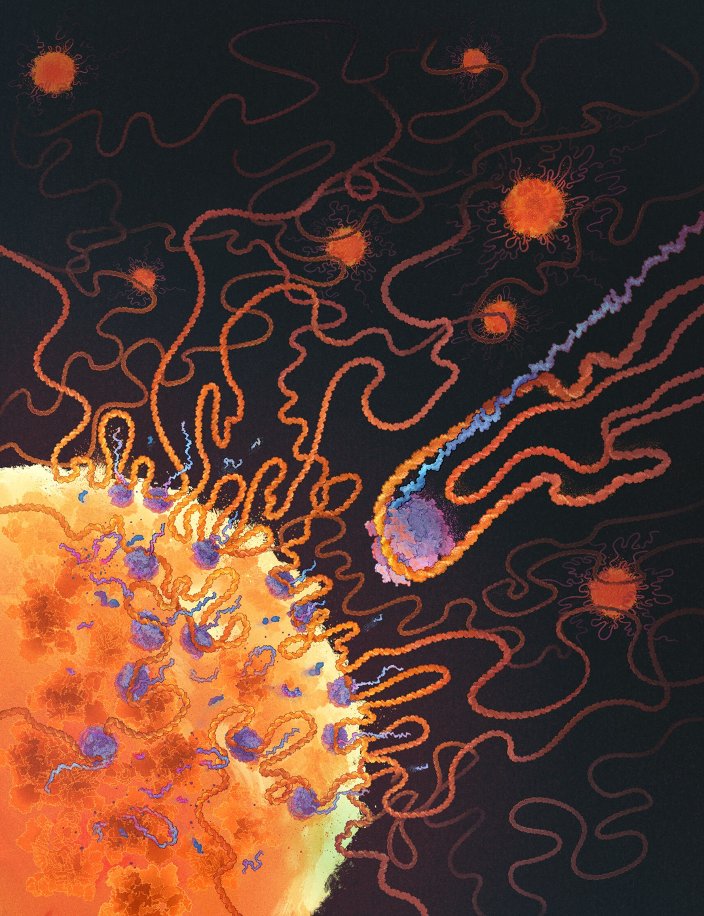
Many proteins bind RNA, yet we still don’t know what RNAs most bind because methods map one RBP at a time. In @CellCellPress, with the Jovanovic lab, we describe SPIDR – a method for mapping the RNA binding sites of dozens of RBPs in a single experiment. sciencedirect.com/science/articl…

Excited that our nucleolus mapping paper just came out today in Nature! Truly an amazing study from even more amazing due of @sofiquinodoz & @jiang_lifei w/ @LafontaineLab & Sebastian Klinge and other fantastic co-authors nature.com/articles/s4158…
Excited to share a new preprint! (1)🔬The nucleolus is the most prominent nuclear condensate, with a fascinating multilayered liquid-like structure, and is the site of ribosome biogenesis. But how does this multiphase architecture form and function? biorxiv.org/content/10.110…

Excited to share our new paper out in @Nature revealing cell-type specific nuclear organization and its link to gene regulation using new spatial multi-omics technologies! nature.com/articles/s4158…
Gene regulation involves thousands of proteins that bind DNA, yet comprehensively mapping these is challenging. Our paper in @NatureGenet describes ChIP-DIP, a method for genome-wide mapping of hundreds of DNA-protein interactions in a single experiment. nature.com/articles/s4158…
Hi! I’ll be speaking at @harvardmed Department of Biomedical Informatics on December 5th about genome organization and splicing/speckles. Thank you to the organizers for the invite! Harvard/MIT/affiliate trainees can RSVP to attend with this link: forms.gle/LopwGCc8RbuCqS…

I am excited to share our review, published online today @NatRevMCB. Pilong Li, Gaofeng Pei, @Heankel_, and I review the current literature on the regulation of transcription by condensates, large length-scale assemblies of complexes (Figure 1). nature.com/articles/s4158…
Delighted to read this article by @TheScientistLLC featuring our work on nuclear speckles! Also features quotes from @mitchguttman and @belmont_andrew. the-scientist.com/gene-proximity…
Elegant study by @ejkk0 & @daSpliceIsRight identifying 5'UTR intron sequences that enhance mRNA expression. Cool to see the integ. reporter + higher Dox leading to greater intron-med. enhancement. Suggests a feedback loop between transcription & splicing to boost gene expression!
Excited to share a new preprint! (1)🔬The nucleolus is the most prominent nuclear condensate, with a fascinating multilayered liquid-like structure, and is the site of ribosome biogenesis. But how does this multiphase architecture form and function? biorxiv.org/content/10.110…

How do you turn a condensate inside-out? Congrats to @sofiquinodoz & @jiang_lifei & co-authors on amazing study from our lab & @LafontaineLab + Klinge. Genomics, engineered nucleoli, & modeling RNA-controlled nucleolar architecture!! (Tweetorial soon!) biorxiv.org/content/10.110…

We are really excited to share this story! We discovered that a major function of the KSHV RNA, kaposin, is as an architectural non-coding RNA that recruits nuclear speckles proximal to viral genomic RNA. biorxiv.org/content/10.110…
Cheer on @UCSFDeptofEM’s Kishan Patel, MD, as he competes on @CBS #Survivor47 in the Fiji islands! This ER doctor thrives in high-pressure situations. Tune in to see how he outwits and outlasts the competition. #UCSFProud @UCSFHealth tiny.ucsf.edu/CvJpUD


I’ve had the chance to interact with all of these amazingly bright and kind humans - congrats to all on their amazing PhDs from @lpachter lab! Go @Caltech!
Proud of newly minted PhDs Tara Chari, @NeuroLuebbert, @tdilan4, and @LambdaMoses (PhD ‘23).

Now Online Splicing regulation through biomolecular condensates and membraneless organelles dlvr.it/T7Bmcw

It was fun to present at my first @RNASociety meeting in Edinburgh! Thanks to the organizers for a great meeting and the audience for the engaging questions! #RNA24 Want to chat about speckles and splicing in different cellular contexts, including disease processes? DM me!

Thanks @philipcball for showcasing our work!
Proud of newly minted PhDs Tara Chari, @NeuroLuebbert, @tdilan4, and @LambdaMoses (PhD ‘23).

Proximity to nuclear speckles–areas dense in mRNA splicing regulators–determines mRNA splicing efficiency. Variation in distance between genes and nuclear speckles drives cell-type specific differences in splicing. @_prashantbhat @mitchguttman ow.ly/cszr50RHV4h

United States Trends
- 1. Wemby 39.1K posts
- 2. Steph 79K posts
- 3. Draymond 17.6K posts
- 4. Spurs 33.8K posts
- 5. Warriors 57.1K posts
- 6. #Truedtac5GXWilliamEst 126K posts
- 7. Clemson 11.3K posts
- 8. Louisville 11K posts
- 9. Zack Ryder 16.9K posts
- 10. Massie 59.5K posts
- 11. PERTHSANTA JOY KAMUTEA 452K posts
- 12. #NEWKAMUEVENTxPerthSanta 447K posts
- 13. #DubNation 2,180 posts
- 14. Bill Clinton 197K posts
- 15. #SmackDown 54.1K posts
- 16. Marjorie Taylor Greene 50.9K posts
- 17. Harden 15.7K posts
- 18. Aaron Fox 2,595 posts
- 19. Bubba 59.9K posts
- 20. Dabo 2,010 posts
You might like
-
 Mitch Guttman
Mitch Guttman
@mitchguttman -
 Lacra Bintu
Lacra Bintu
@BintuLacra -
 brangwynnelab
brangwynnelab
@brangwynnelab -
 Whitney Griggs
Whitney Griggs
@WhitneySGriggs -
 Miguel (Miggy) Chuapoco
Miguel (Miggy) Chuapoco
@MiggyChuapoco -
 Machine learning for protein engineering seminar
Machine learning for protein engineering seminar
@ml4proteins -
 ASHansen Lab
ASHansen Lab
@hansen_lab -
 LomvardasLab
LomvardasLab
@LomvardasLab -
 Nature Conferences
Nature Conferences
@NatureConf -
 CERNpress
CERNpress
@CERNpress -
 Sam Sternberg
Sam Sternberg
@shsternberg -
 Delaney Sullivan
Delaney Sullivan
@DelaneyKSull
Something went wrong.
Something went wrong.