#noncodingvariants результаты поиска
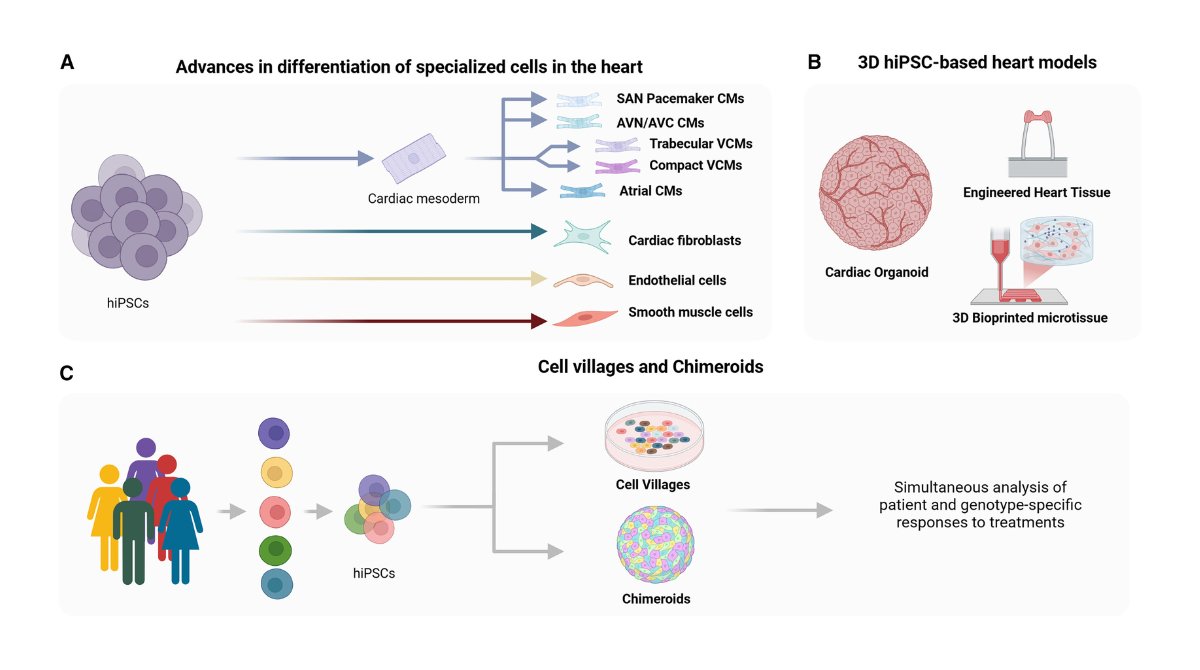
Online now! Review discussing the basics of noncoding variant-mediated pathogenesis, the methodologies utilized, and how #iPSC -based heart models can be leveraged to dissect the mechanisms of #noncodingvariants ow.ly/wqcH50VlLGC @UBCcps @BCCHresearch @SBME_UBC @ISSCR
Classification of non-coding variants with high pathogenic impact. #WGS #NonCodingVariants #FunctionalPrediction #DiseaseImpact #MachineLearning #Bioinformatics journals.plos.org/plosgenetics/a… @PLOSGenetics
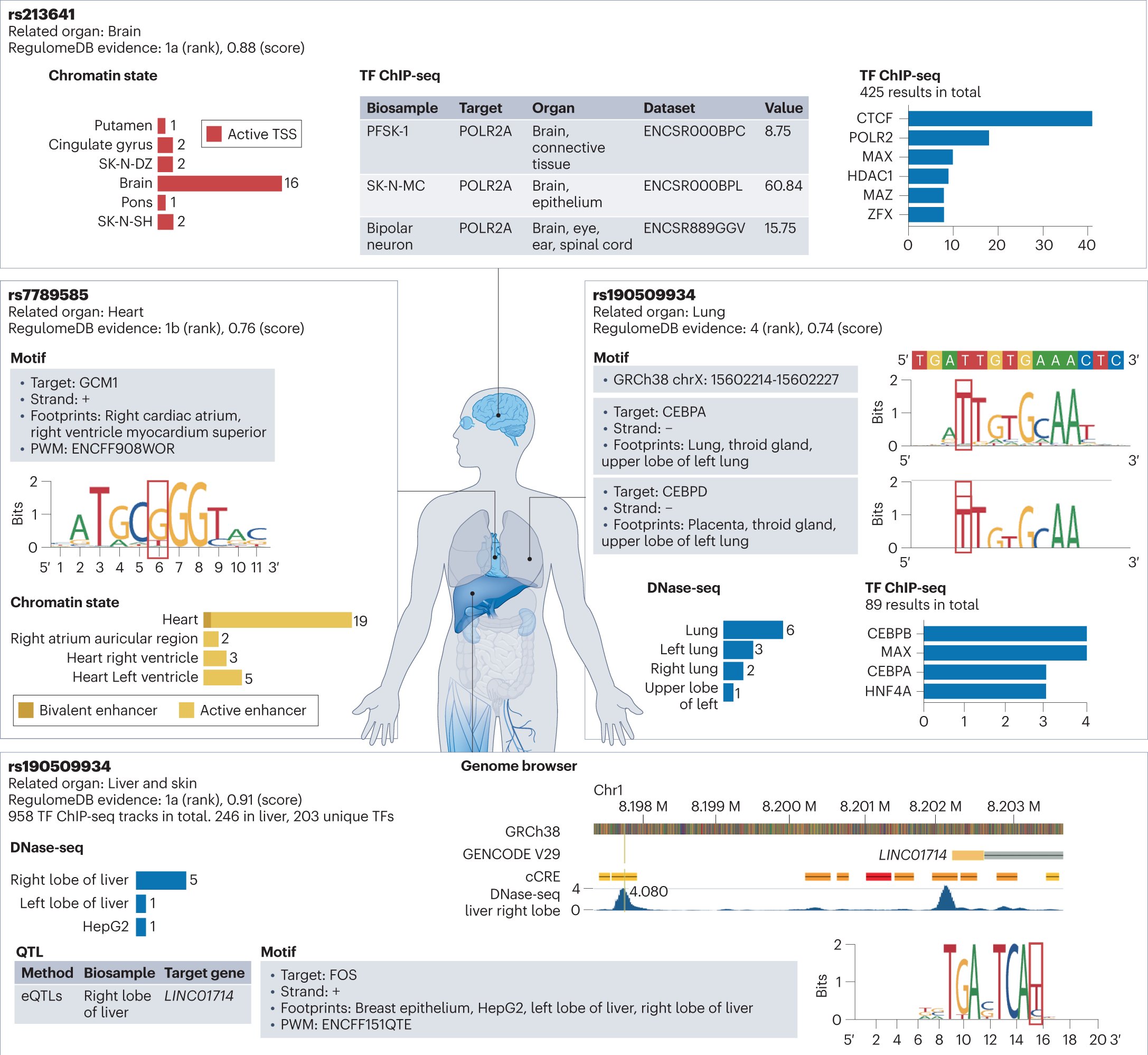
Annotating and prioritizing human non-coding variants with RegulomeDB v.2. #NonCodingVariants #VariantsAnnotation #VariantsPrioritization @NatureGenet nature.com/articles/s4158…
Recommendations for clinical interpretation of variants found in non-coding regions of the genome. #NonCodingVariants #Clinicalinterpretation @j_ellingford @nickywhiffin @GenomeMedicine genomemedicine.biomedcentral.com/articles/10.11…
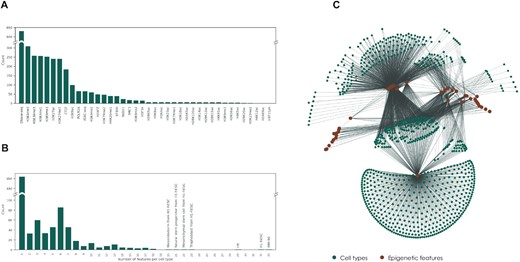
Cell type–specific interpretation of noncoding variants using deep learning–based methods. #NonCodingVariants #DeepLearning #CellTypeVariants @GigaScience academic.oup.com/gigascience/ar…
New Topic "#NonCodingVariants: From #Interpretation to #PersonalizedMedicine" led by Drs Elvezia Maria Paraboschi @HumanitasMilano, Elisabetta Morini and Harrison Brand @Harvard and @hongheeW is open for submissions. Submit here: fro.ntiers.in/3oa7

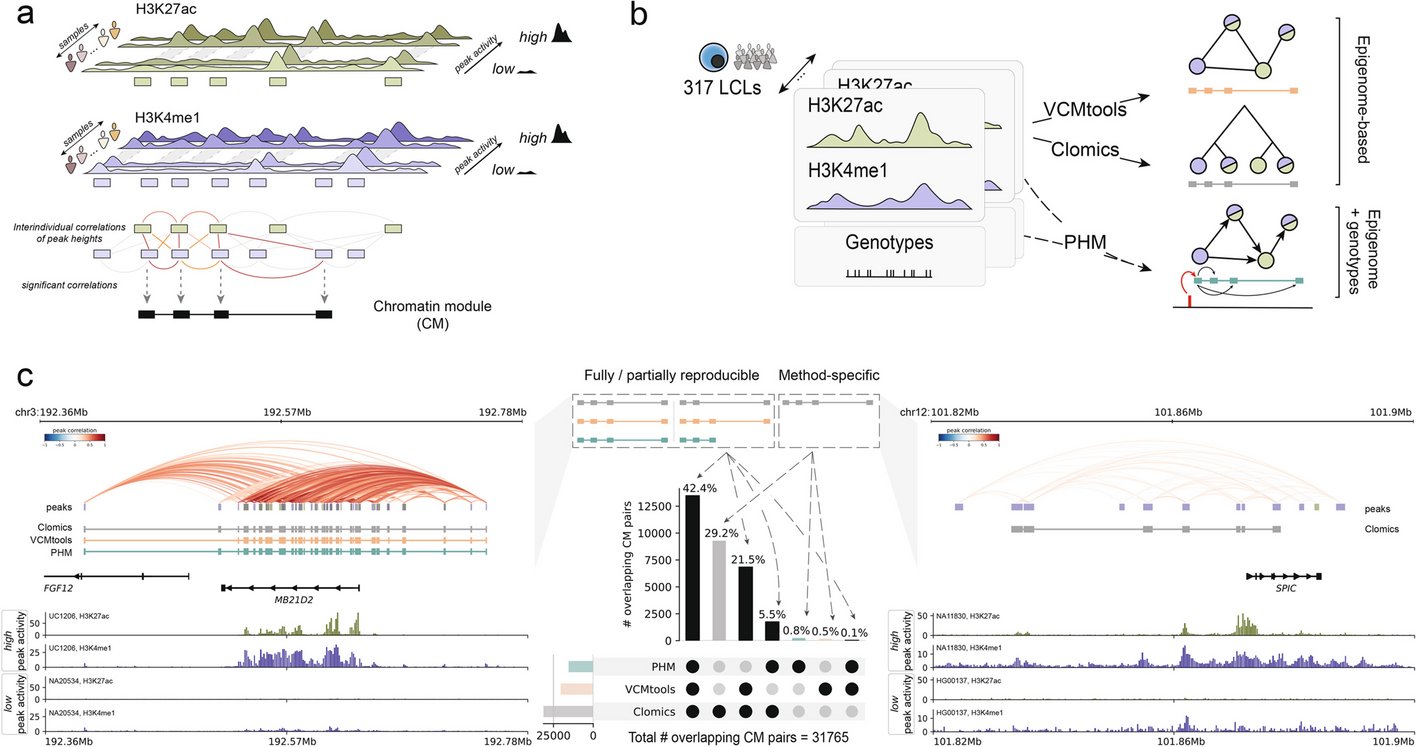
Non-coding variants impact cis-regulatory coordination in a cell type-specific manner. #NonCodingVariants #CisRegulatoryElements @GenomeBiology genomebiology.biomedcentral.com/articles/10.11…
“By using aberrant #genetranscription to reveal the function of #noncodingvariants, we developed cis-X to enable discovery of noncoding variants driving #cancer in individual tumor #genomes" - @StJudeResearch rna-seqblog.com/st-jude-resear…
Classification of non-coding variants with high pathogenic impact. #NonCodingVariants #PathogenicityPrioritization journals.plos.org/plosgenetics/a… @PLOSGenetics
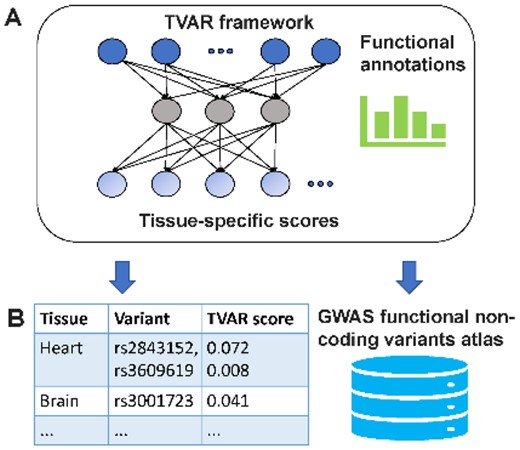
TVAR: Assessing Tissue-specific Functional Effects of Non-coding Variants with Deep Learning. #NonCodingVariants #DeepLearning #Bioinformatics academic.oup.com/bioinformatics…
A framework for detecting noncoding rare variant associations of large-scale whole-genome sequencing studies. #NonCodingVariants #WGSdata #Bioinformatics biorxiv.org/content/10.110…
Performance Comparison of Computational Prediction Methods for the Function and Pathogenicity of Non-coding Variants. #NonCodingVariants #FunctionPrediction #ToolsBenchmarking biorxiv.org/content/10.110… @biorxivpreprint #Bioinformatics
Structural and non-coding variants increase the diagnostic yield of clinical whole genome sequencing for rare diseases. #StructuralVariants #SVs #NoncodingVariants #Diagnostic #WGS #RareDiseases @GenomeMedicine genomemedicine.biomedcentral.com/articles/10.11…
WEVar: a novel statistical learning framework for predicting noncoding regulatory variants. #NonCodingVariants academic.oup.com/bib/advance-ar… @BriefingBioinfo
Great talk from @j_ellingford on interpreting #NonCodingVariants in @GenomicsEngland data with focus in ophthalmological disorders #LGN2022 @LdnGeneNet
The Impact of Rare Noncoding Variants scienceness.com/news/6667-the-… #Scienceness #TranscriptomeSequencing #NoncodingVariants #eQTLs #sQTLs
Reclutada durante años en Dpto. Genética @Hospital_FJD, el Servicio de Oftalmología del HU Clínico San Carlos y Fac Med Albacete. Esperamos además mejorar las tasas de caracterización genética 🧬, identificar nuevos genes, implicación #noncodingvariants 🧵
“Predicting the impact of #noncodingvariants is still a challenge” @alexisjbattle #WGS & #RNA-seq data GTEx #bog16 genomeweb.com/sequencing-tec…
#BankaGrpJC 03_2020 Journal Club on #Zoom! Last week we discussed this interesting article on #NoncodingVariants #SNPs rdcu.be/b3qej
Great talk from @j_ellingford on interpreting #NonCodingVariants in #RareDisease 🧬 Highlighted available New guidelines for standardized clinical variant interpretation & that Functional information & methods used are critical for variant interpretation. #LGN2022 @LdnGeneNet
Online now! Review discussing the basics of noncoding variant-mediated pathogenesis, the methodologies utilized, and how #iPSC -based heart models can be leveraged to dissect the mechanisms of #noncodingvariants ow.ly/wqcH50VlLGC @UBCcps @BCCHresearch @SBME_UBC @ISSCR

Reclutada durante años en Dpto. Genética @Hospital_FJD, el Servicio de Oftalmología del HU Clínico San Carlos y Fac Med Albacete. Esperamos además mejorar las tasas de caracterización genética 🧬, identificar nuevos genes, implicación #noncodingvariants 🧵
Non-coding variants impact cis-regulatory coordination in a cell type-specific manner. #NonCodingVariants #CisRegulatoryElements @GenomeBiology genomebiology.biomedcentral.com/articles/10.11…
Structural and non-coding variants increase the diagnostic yield of clinical whole genome sequencing for rare diseases. #StructuralVariants #SVs #NoncodingVariants #Diagnostic #WGS #RareDiseases @GenomeMedicine genomemedicine.biomedcentral.com/articles/10.11…
Annotating and prioritizing human non-coding variants with RegulomeDB v.2. #NonCodingVariants #VariantsAnnotation #VariantsPrioritization @NatureGenet nature.com/articles/s4158…
Cell type–specific interpretation of noncoding variants using deep learning–based methods. #NonCodingVariants #DeepLearning #CellTypeVariants @GigaScience academic.oup.com/gigascience/ar…
Great talk from @j_ellingford on interpreting #NonCodingVariants in @GenomicsEngland data with focus in ophthalmological disorders #LGN2022 @LdnGeneNet
Great talk from @j_ellingford on interpreting #NonCodingVariants in #RareDisease 🧬 Highlighted available New guidelines for standardized clinical variant interpretation & that Functional information & methods used are critical for variant interpretation. #LGN2022 @LdnGeneNet
TVAR: Assessing Tissue-specific Functional Effects of Non-coding Variants with Deep Learning. #NonCodingVariants #DeepLearning #Bioinformatics academic.oup.com/bioinformatics…
Recommendations for clinical interpretation of variants found in non-coding regions of the genome. #NonCodingVariants #Clinicalinterpretation @j_ellingford @nickywhiffin @GenomeMedicine genomemedicine.biomedcentral.com/articles/10.11…
Classification of non-coding variants with high pathogenic impact. #WGS #NonCodingVariants #FunctionalPrediction #DiseaseImpact #MachineLearning #Bioinformatics journals.plos.org/plosgenetics/a… @PLOSGenetics
Classification of non-coding variants with high pathogenic impact. #NonCodingVariants #PathogenicityPrioritization journals.plos.org/plosgenetics/a… @PLOSGenetics
A framework for detecting noncoding rare variant associations of large-scale whole-genome sequencing studies. #NonCodingVariants #WGSdata #Bioinformatics biorxiv.org/content/10.110…
Performance Comparison of Computational Prediction Methods for the Function and Pathogenicity of Non-coding Variants. #NonCodingVariants #FunctionPrediction #ToolsBenchmarking biorxiv.org/content/10.110… @biorxivpreprint #Bioinformatics
WEVar: a novel statistical learning framework for predicting noncoding regulatory variants. #NonCodingVariants academic.oup.com/bib/advance-ar… @BriefingBioinfo
Classification of non-coding variants with high pathogenic impact. #WGS #NonCodingVariants #FunctionalPrediction #DiseaseImpact #MachineLearning #Bioinformatics biorxiv.org/content/10.110…
New Topic "#NonCodingVariants: From #Interpretation to #PersonalizedMedicine" led by Drs Elvezia Maria Paraboschi @HumanitasMilano, Elisabetta Morini and Harrison Brand @Harvard and @hongheeW is open for submissions. Submit here: fro.ntiers.in/3oa7

“By using aberrant #genetranscription to reveal the function of #noncodingvariants, we developed cis-X to enable discovery of noncoding variants driving #cancer in individual tumor #genomes" - @StJudeResearch rna-seqblog.com/st-jude-resear…
#BankaGrpJC 03_2020 Journal Club on #Zoom! Last week we discussed this interesting article on #NoncodingVariants #SNPs rdcu.be/b3qej
“Predicting the impact of #noncodingvariants is still a challenge” @alexisjbattle #WGS & #RNA-seq data GTEx #bog16 genomeweb.com/sequencing-tec…
Online now! Review discussing the basics of noncoding variant-mediated pathogenesis, the methodologies utilized, and how #iPSC -based heart models can be leveraged to dissect the mechanisms of #noncodingvariants ow.ly/wqcH50VlLGC @UBCcps @BCCHresearch @SBME_UBC @ISSCR

New Topic "#NonCodingVariants: From #Interpretation to #PersonalizedMedicine" led by Drs Elvezia Maria Paraboschi @HumanitasMilano, Elisabetta Morini and Harrison Brand @Harvard and @hongheeW is open for submissions. Submit here: fro.ntiers.in/3oa7

Something went wrong.
Something went wrong.
United States Trends
- 1. Broncos 46.2K posts
- 2. Mariota 12.9K posts
- 3. Ertz 3,080 posts
- 4. Commanders 32.9K posts
- 5. Bo Nix 9,899 posts
- 6. #RaiseHail 5,714 posts
- 7. Riley Moss 2,266 posts
- 8. #BaddiesUSA 25.1K posts
- 9. Bobby Wagner 1,045 posts
- 10. Burks 15.4K posts
- 11. Terry 20.4K posts
- 12. #DENvsWAS 3,169 posts
- 13. Happy New Month 156K posts
- 14. Bonitto 5,621 posts
- 15. Deebo 3,105 posts
- 16. Collinsworth 3,033 posts
- 17. #RHOP 12.2K posts
- 18. Dan Quinn N/A
- 19. Sean Payton 1,615 posts
- 20. Zach Edey 3,048 posts