#moleculardynamics ผลการค้นหา
1/ 🚀 BIG NEWS in the world of biology & molecular simulations! The Molecular dynamics simulation (MDS) is about to get a major upgrade. But first, let's talk about what it is and why it matters. #MolecularDynamics #compchem #structuralbiology #protein 👇

1⃣ Featuring the first of our showcase projects We use an autoencoder to capture and quantify dynamic differences between protein conformational states ➡️ bioexcel.eu/wv4a #MolecularDynamics #MachineLearning

🚀 awesome-AI4MolConformation-MD Explore cutting-edge tools for modeling molecular conformations and dynamics using generative AI and deep learning! covering MD engines, force fields, neural potentials, and ensemble predictions. github.com/AspirinCode/aw… #MolecularDynamics #AI4MD

#moleculardynamics (MD), #CADD, #quantummechanics (QM), #artificialintelligence, #ChemicalSpace, Knowledge Bases, #Freeenergy, #AI, #insilico #ADMET, #Molecularglues, and Computing Technologies such as #cloudcomputing and #quantumcomputing in one book

Computational drug discovery book #moleculardynamics #qm #NMR #comchem #drugdiscovery #wiley #chemicalspace #QuantumComputing #CloudComputing wiley-vch.de/en?option=com_…

🧵1⃣ Scientists introduce #Dynaformer, a revolutionary graph-based #DeepLearning model for predicting protein-ligand binding affinities. It leverages #moleculardynamics simulations to capture the dynamic nature of protein-ligand interactions. Quick Read: cbirt.net/unlocking-mole…

On-the-Fly Sequential Design of Simple Peptides #MolecularDynamics pubs.acs.org/doi/10.1021/ac… @F_Coppola93 #JCIM Vol65 Issue18 #PharmaceuticalModeling
2⃣ Featuring the second of our showcase projects Through a combination of AlphaFold2 and other tools we are bypassing the limits of traditional MD simulations ➡️ bioexcel.eu/dvy3 #Proteins #MolecularDynamics #GROMACS #AlphaFold2

Molecular Dynamics Simulations Reveal Conformational Determinants of the Dynamic Association between α-Synuclein and Membranes #MolecularDynamics pubs.acs.org/doi/10.1021/ac… #JCIM Vol65 Issue18 #compchem
Binding of Antimicrobial Peptide Indolicidin to DMPC Bilayer Using Replica-Exchange Molecular Dynamics #MolecularDynamics pubs.acs.org/doi/10.1021/ac… #JCIM Vol65 Issue17 #compchem
🌟 Thread Alert! Dive into NPT Equilibration using GROMACS Wizard. Master this essential step in molecular dynamics with SAMSON! First, let's set up the simulation. #MolecularDynamics #GROMACS

Exploring Peptide’s Antifreeze Activity Using a Semi-Automated Molecular Dynamics-Enabled Screening Framework #MolecularDynamics pubs.acs.org/doi/10.1021/ac… #JCIM Vol65 Issue19 #ApplicationNote
Efficient Free Energies from a Simplified Electrostatic Embedding QM/MM Approach Based on Electrostatic Potential Fitted Operators #MolecularDynamics #DFT pubs.acs.org/doi/10.1021/ac… @HuixRotllant #JCIM Vol65 Issue17 #compchem
Conformational Ensemble Dynamics of Intrinsically Disordered Full-Length α- and β-Synuclein Monomers #MolecularDynamics pubs.acs.org/doi/10.1021/ac… #JCIM Vol65 Issue17 #compchem
Decoding BCL6 Inhibitors: Computational Insights into the Impact of Water Networks on Potency #MolecularDynamics pubs.acs.org/doi/10.1021/ac… @andrea_scarpino @TheRealB84 @ICR_London #JCIM Vol65 Issue18 #compchem
Solvation Energetic Costs of Cognate Binding Site Formation #MolecularDynamics pubs.acs.org/doi/10.1021/ac… @KurtzmanTom #JCIM Vol65 Issue17 #compchem
From AI-Driven Sequence Generation to Molecular Simulation: A Comprehensive Framework for Antimicrobial Peptide Discovery #MolecularDynamics pubs.acs.org/doi/10.1021/ac… #JCIM Vol65 Issue18 #compchem
A Practical Covariance-Based Method for Efficient Detection of Protein–Protein Attractive and Repulsive Interactions in Molecular Dynamics Simulations #MolecularDynamics pubs.acs.org/doi/10.1021/ac… @PittGurLab #JCIM Vol65 Issue19 #Letter
Conserved Protonation Pattern in the Extended Active Site of Human Carboxylesterase 1 and Its Impact on Enzyme Catalysis #MolecularDynamics pubs.acs.org/doi/10.1021/ac… #JCIM Vol65 Issue16 #compchem
𝗦𝗧𝗔𝗥𝗧𝗜𝗡𝗚 𝗜𝗡 𝟮 𝗛𝗢𝗨𝗥𝗦. .𝗟𝗔𝗦𝗧 𝗖𝗛𝗔𝗡𝗖𝗘 𝗧𝗢 𝗥𝗘𝗚𝗜𝗦𝗧𝗘𝗥 🔗 Register now: lnkd.in/gebjvenM #MolecularDynamics #GROMACS #Bioinformatics #ComputationalBiology #ProteinSimulation #TIBRBiotech
𝐒𝐭𝐚𝐫𝐭𝐢𝐧𝐠 𝐓𝐎𝐃𝐀𝐘. 𝐅𝐢𝐧𝐚𝐥 𝐂𝐡𝐚𝐧𝐜𝐞 𝐭𝐨 𝐑𝐄𝐆𝐈𝐒𝐓𝐄𝐑. . 🧬 LIVE Hands-on Workshop on MOLECULAR DYNAMICS SIMULATION with GROMACS 💻 🔗 Register now: lnkd.in/gebjvenM #MolecularDynamics #GROMACS #Bioinformatics #ComputationalBiology #tibrbiotech
𝐖𝐎𝐑𝐊𝐒𝐇𝐎𝐏 𝐁𝐄𝐆𝐈𝐍𝐒 𝐓𝐎𝐌𝐎𝐑𝐑𝐎𝐖. . 𝐑𝐄𝐆𝐈𝐒𝐓𝐄𝐑 𝐍𝐎𝐖... 🧬 LIVE Hands-on Workshop on MOLECULAR DYNAMICS SIMULATION with GROMACS 💻 🔗 Register now: lnkd.in/gebjvenM #MolecularDynamics #GROMACS #Bioinformatics #ComputationalBiology #TIBRBiotech
𝐖𝐎𝐑𝐊𝐒𝐇𝐎𝐏 𝐁𝐄𝐆𝐈𝐍𝐒 𝐓𝐎𝐌𝐎𝐑𝐑𝐎𝐖. . 𝐑𝐄𝐆𝐈𝐒𝐓𝐄𝐑 𝐍𝐎𝐖... 🧬 LIVE Hands-on Workshop on MOLECULAR DYNAMICS SIMULATION with GROMACS 💻 🔗 Register now: lnkd.in/gebjvenM #MolecularDynamics #GROMACS #Bioinformatics #ComputationalBiology #TIBRBiotech
𝐖𝐎𝐑𝐊𝐒𝐇𝐎𝐏 𝐁𝐄𝐆𝐈𝐍𝐒 𝐓𝐎𝐌𝐎𝐑𝐑𝐎𝐖. . 𝐑𝐄𝐆𝐈𝐒𝐓𝐄𝐑 𝐍𝐎𝐖... 🧬 LIVE Hands-on Workshop on MOLECULAR DYNAMICS SIMULATION with GROMACS 💻 🔗 Register now: lnkd.in/gebjvenM #MolecularDynamics #GROMACS #Bioinformatics #ComputationalBiology #TIBRBiotech

𝗦𝗧𝗔𝗥𝗧𝗜𝗡𝗚 𝗜𝗡 𝟮 𝗗𝗔𝗬𝗦. 𝗥𝗘𝗚𝗜𝗦𝗧𝗘𝗥 𝗡𝗢𝗪 ..𝗢𝗡𝗟𝗬 𝗟𝗜𝗠𝗜𝗧𝗘𝗗 𝗦𝗘𝗔𝗧𝗦 𝗟𝗘𝗙𝗧.. 🧬 LIVE Hands-on Workshop on MOLECULAR DYNAMICS SIMULATION with GROMACS 💻 🔗 Register now: lnkd.in/gebjvenM #MolecularDynamics
𝗦𝗧𝗔𝗥𝗧𝗜𝗡𝗚 𝗜𝗡 𝟮 𝗗𝗔𝗬𝗦. 𝗥𝗘𝗚𝗜𝗦𝗧𝗘𝗥 𝗡𝗢𝗪 ..𝗢𝗡𝗟𝗬 𝗟𝗜𝗠𝗜𝗧𝗘𝗗 𝗦𝗘𝗔𝗧𝗦 𝗟𝗘𝗙𝗧.. 🧬 LIVE Hands-on Workshop on MOLECULAR DYNAMICS SIMULATION with GROMACS 💻 🔗 Register now: lnkd.in/gebjvenM #MolecularDynamics #GROMACS #Bioinformatics #TIBRBiotech

A Practical Covariance-Based Method for Efficient Detection of Protein–Protein Attractive and Repulsive Interactions in Molecular Dynamics Simulations #MolecularDynamics pubs.acs.org/doi/10.1021/ac… @PittGurLab #JCIM Vol65 Issue19 #Letter
🧬 LIVE Hands-on Workshop on MOLECULAR DYNAMICS SIMULATION with GROMACS 💻 📅 Oct 30 – Nov 2, 2025 🕖 7:00 – 8:30 PM IST 🎥 Video recordings shared daily 📜 Course completion certificate provided 🔗 Register now: lnkd.in/gebjvenM #MolecularDynamics #GROMACS

🧬 LIVE Hands-on Workshop on MOLECULAR DYNAMICS SIMULATION with GROMACS 💻 📅 Oct 30 – Nov 2, 2025 🕖 7:00 – 8:30 PM IST 🎥 Video recordings shared daily 📜 Course completion certificate provided 🔗 Register now: lnkd.in/gebjvenM #MolecularDynamics
🧬 LIVE Hands-on Workshop on MOLECULAR DYNAMICS SIMULATION with GROMACS 💻 📅 Oct 30 – Nov 2, 2025 🔗 Register now: lnkd.in/gebjvenM #MolecularDynamics #GROMACS #Bioinformatics #ComputationalBiology #ProteinSimulation #TIBRBiotech #OnlineWorkshop #StructuralBiology
Applications are invited for the position of Project Associate–I under the DHR Start-Up Grant project entitled: “Investigating the Role of p53 in Post-Mortem Interval (PMI) Estimation Using Molecular Dynamics Simulations.” #Bioinformatics #MolecularDynamics #ICMR #DHR #RGU #FSc

Applications are invited for the position of Project Associate–I under the DHR Start-Up Grant project entitled: “Investigating the Role of p53 in Post-Mortem Interval (PMI) Estimation Using Molecular Dynamics Simulations.” #Bioinformatics #MolecularDynamics #ICMR #DHR #RGU #FSc

Molecular dynamics isn’t just a science, it’s motion at the atomic level. Here 4 protein–peptide complexes are shaking, flexing, adapting. Expensive to compute 💻, priceless for discovery 💡. This is how I see peptides… #MolecularDynamics #ComputationalChemistry #DrugDesign
1/ 🚀 BIG NEWS in the world of biology & molecular simulations! The Molecular dynamics simulation (MDS) is about to get a major upgrade. But first, let's talk about what it is and why it matters. #MolecularDynamics #compchem #structuralbiology #protein 👇

1⃣ Featuring the first of our showcase projects We use an autoencoder to capture and quantify dynamic differences between protein conformational states ➡️ bioexcel.eu/wv4a #MolecularDynamics #MachineLearning

2⃣ Featuring the second of our showcase projects Through a combination of AlphaFold2 and other tools we are bypassing the limits of traditional MD simulations ➡️ bioexcel.eu/dvy3 #Proteins #MolecularDynamics #GROMACS #AlphaFold2

Our very own Thomas Tarenzi presents a fascinating talk on fuzzy interactions on proteins in the yeast acetylation complex at the frontiers in IDP meeting #IDP #moleculardynamics

🧵1⃣ Scientists introduce #Dynaformer, a revolutionary graph-based #DeepLearning model for predicting protein-ligand binding affinities. It leverages #moleculardynamics simulations to capture the dynamic nature of protein-ligand interactions. Quick Read: cbirt.net/unlocking-mole…

4⃣ Featuring the fourth of our showcase projects Upgrading GROMACS to handle billion-atom systems and enhancing I/O performance and precision, making the first-ever whole-cell simulation possible ➡️ bioexcel.eu/uw67 #MolecularDynamics #GROMACS #ComputationalBiology

Excited for #BPS2023! All @palermo_lab group members will join the conference! Do not miss their posters/talks if you want to know more about our work on #moleculardynamics, #compchem, #genomeediting, #CRISPR, #RNA and other exciting topics in Biophysics!

📢#Publicationalert Work from Dr. P.B. Raghavendra's lab reveals interaction of DPP4 with SARS-CoV-2 variants and MERS-CoV which will help optimize therapeutic use of DPP4 to manage COVID-19 disease severity. Read more: bit.ly/3QbL3eM #imunoinformatics #moleculardynamics

❇️Differences between the GluD1 & GluD2 receptors revealed by X-ray #crystallography, binding studies & #moleculardynamics❇️ ✏️By Kastrup, Jørgensen, Hollmann et al @UCPH_Research @ruhrunibochum ➡️Article: buff.ly/3QkVQUj Commentary: buff.ly/4496aCd #OpenAccess

📚 Structural dynamics of the N-terminal SH2 domain of PI3K in its free and CD28-bound states ✏️ By Masayuki Oda from @osaka_univ_e @mcgc_jp 🔗 buff.ly/3plTSaH #conformationalchange #moleculardynamics #NMR

🔬💻 A new tool in the toolbox: in a recent Review article, Jennifer Sapia and Stefano Vanni discuss how MD simulations have enhanced our understanding of lipid droplets doi.org/10.1002/1873-3… #lipiddroplets #MolecularDynamics @jennifersapia17

Calcium binding and permeation in TRPV channels: Insights from #MolecularDynamics simulations. A new study from Chunhong Liu, Lingfeng Xue, and Chen Song @PKU1898: bit.ly/3RrYMyO #Biophysics #IonChannels

[Selected Paper] #ProteinProteinInteractions , #MolecularDynamics simulation , #AntitumorAgents Article by Prof. Masaki Kita @NagoyaUniv (Nagoya University) #ProteinProteinInteraction #MDsimulation #OnTheCover #FreeAccess academic.oup.com/bcsj/article/9…
![CSJjournals_jp's tweet image. [Selected Paper]
#ProteinProteinInteractions , #MolecularDynamics simulation , #AntitumorAgents
Article by Prof. Masaki Kita @NagoyaUniv (Nagoya University)
#ProteinProteinInteraction #MDsimulation #OnTheCover #FreeAccess
academic.oup.com/bcsj/article/9…](https://pbs.twimg.com/media/GWr3wRGa8AAMzGK.jpg)
🌟 Thread Alert! Dive into NPT Equilibration using GROMACS Wizard. Master this essential step in molecular dynamics with SAMSON! First, let's set up the simulation. #MolecularDynamics #GROMACS

Sriraksha Srinivasan, Daniel Álvarez, Stefano Vanni et al. @LabVanni @unifr develop a fast and easy-to-use protocol based on coarse-grained #moleculardynamics simulations to characterize #lipid binding to lipid transfer proteins. hubs.la/Q02KfsXq0 #lipids #biophysics

Woops... I guess something has gone wrong. :) #structuralbiology #mdsimulation #moleculardynamics #openmm #bioinformatics
#moleculardynamics (MD), #CADD, #quantummechanics (QM), #artificialintelligence, #ChemicalSpace, Knowledge Bases, #Freeenergy, #AI, #insilico #ADMET, #Molecularglues, and Computing Technologies such as #cloudcomputing and #quantumcomputing in one book

Congrats to Cianna, who passed her 2nd-year exam with flying colors today! On the road from #ICU to #PhD, she's engineering antifreeze proteins with #MolecularDynamics and #MachineLearning. A true & inspiring #pirate! Join #TeamCianna 👉 gofund.me/b04f2591! #phdlife
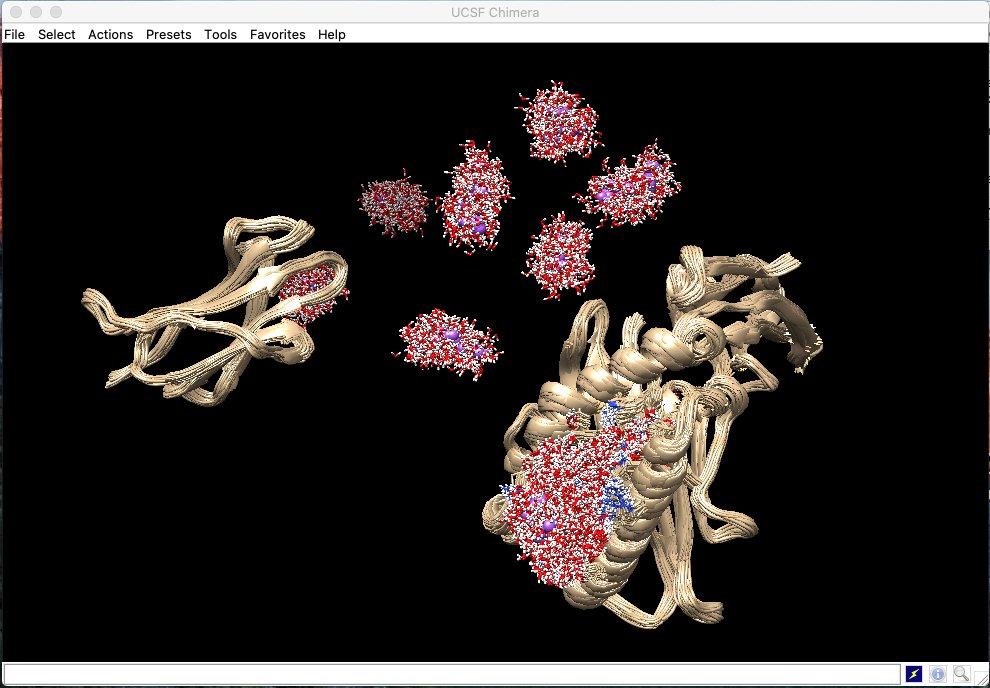
🚀 Exciting new poster from our lab at #BPS2025! 🧬 @Ahsan09966804 showcases how #MolecularDynamics & #AI can enhance #CryoEM map refinement. 📍 Catch it at Board 18 (1824-Pos) today, don’t miss out! #Biophysics #CompBio

Espinoza-Arcos, González-Avendaño, Vergara-Jaque, Poblete et al. @UTalca identify a peripheral PIP2-binding site conserved across TRP channels using coarse-grained #MolecularDynamics simulations. hubs.la/Q03F10LN0 #Lipids #ComputationalBiology #IonChannels

Something went wrong.
Something went wrong.
United States Trends
- 1. #warmertogether N/A
- 2. $BARRON 2,034 posts
- 3. Harvey Weinstein 2,841 posts
- 4. Diane Ladd 3,004 posts
- 5. Ben Shapiro 27K posts
- 6. $PLTR 16.5K posts
- 7. #NXXT 2,505 posts
- 8. Laura Dern 1,413 posts
- 9. Gold's Gym 48K posts
- 10. #maddiekowalski 3,894 posts
- 11. #CAVoteYesProp50 5,515 posts
- 12. Cardinals 11.8K posts
- 13. iOS 26.1 3,142 posts
- 14. Standout 7,836 posts
- 15. University of Virginia 1,802 posts
- 16. Mumdumi 12.2K posts
- 17. $HIMS 6,068 posts
- 18. #BestStockToBuy 1,116 posts
- 19. Shannon Library 1,830 posts
- 20. McBride 2,950 posts