#spatial_multiomics résultats de recherche
Postdoctoral position in spatial transcriptomics Postdoctoral position in spatial transcriptomics at Karolinska Institutet jobrxiv.org/job/karolinska… #bioinformatics #spatial_multiomics #spatial_transcriptomics #ScienceJobs #job
Postdoctoral position in spatial transcriptomics Postdoctoral position in spatial transcriptomics at Karolinska Institutet jobrxiv.org/job/karolinska… #bioinformatics #spatial_multiomics #spatial_transcriptomics #ScienceJobs #job
Postdoctoral position in spatial transcriptomics Postdoctoral position in spatial transcriptomics at Karolinska Institutet jobrxiv.org/job/karolinska… #bioinformatics #spatial_multiomics #spatial_transcriptomics #ScienceJobs #job
MultiModalGraphics: an R package for graphical integration of multi-omics datasets bmcbioinformatics.biomedcentral.com/articles/10.11…
How close can science get to reality? Multiomics is more than a method – it’s a shift in perception. By observing DNA, RNA, proteins, and metabolites within the same single cell, we move from fragments to a connected whole. In our latest SelectScience article, we show how…

Announcing our Frontier Data Centers Hub! The world is about to see multiple 1 GW+ AI data centers. We mapped their construction using satellite imagery, permits & public sources — releasing everything for free, including commissioned satellite images. Highlights in thread!

Later today you can discover how the computational analysis of high-plex imaging can enhance spatial resolution and reproducibility to advance biological insights? Sign-up here! >> hubs.ly/Q03PtCp70

3 amazing papers just out @Nature, the kind worth giving up sleep for🦉 Spatial multi-omics human maps: -placenta: MIBI & DSP -intestine: CODEX & snRNAseq & snATACseq -kidney: Visium & scRNA & scATAC After sequencing single cells, we are now finally putting them back together🧵

[PUBLI] EMito-Metrix, a novel tool for mitochondria analysis A team from the Restore Institute has developed EMito-Metrix, a computational tool designed for the automatic segmentation and analysis of mitochondria from 2D EM images. 🔗 Read our article: bit.ly/495cwZg
![Fr_BioImaging's tweet image. [PUBLI] EMito-Metrix, a novel tool for mitochondria analysis
A team from the Restore Institute has developed EMito-Metrix, a computational tool designed for the automatic segmentation and analysis of mitochondria from 2D EM images.
🔗 Read our article: bit.ly/495cwZg](https://pbs.twimg.com/media/G5FXP2oWkAAPvEX.jpg)
New open access article online: Multispectral live-cell imaging with uncompromised spatiotemporal resolution. go.nature.com/3JA4DR6

CellSP enables module discovery and visualization for subcellular spatial transcriptomics data dlvr.it/TP5fkb #MachineLearning #NatureJournal #AI

🚨New #SpatialTranscriptomics #Bioinformatics data resource out in @naturemethods. SODB, a platform with >2,400 manually curated spatial experiments from >25 spatial omics technologies & interactive analytical modules. This🧵will walk you through all the features of SODB [1/33]
![simocristea's tweet image. 🚨New #SpatialTranscriptomics #Bioinformatics data resource out in @naturemethods.
SODB, a platform with >2,400 manually curated spatial experiments from >25 spatial omics technologies & interactive analytical modules.
This🧵will walk you through all the features of SODB [1/33]](https://pbs.twimg.com/media/FpqfK6HaUAAQZAo.jpg)
the schematics, and the actual setup ( multispectral live-cell imaging from Cambridge )


🔍 Data meets geography! 📖 Explore the studies ↓ 1️⃣ bmjpublichealth.bmj.com/content/3/2/e0… 2️⃣ link.springer.com/article/10.100… @ICMRDELHI @DeptHealthRes #DigitalHealth #GIS #SpatialAnalysis #WeAreICMR

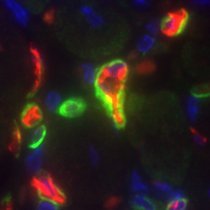
Check this image, it's a single picture right? Nope! It's 120 images combined, 36 µm of tissue. Acquiring data in 3D will allow you to expand your analysis, and for example see how neurons and blood vessels interact #3DThursday 🟡Neuron 🔵Astrocyte 🟣Microglia 🔴Red blood cell

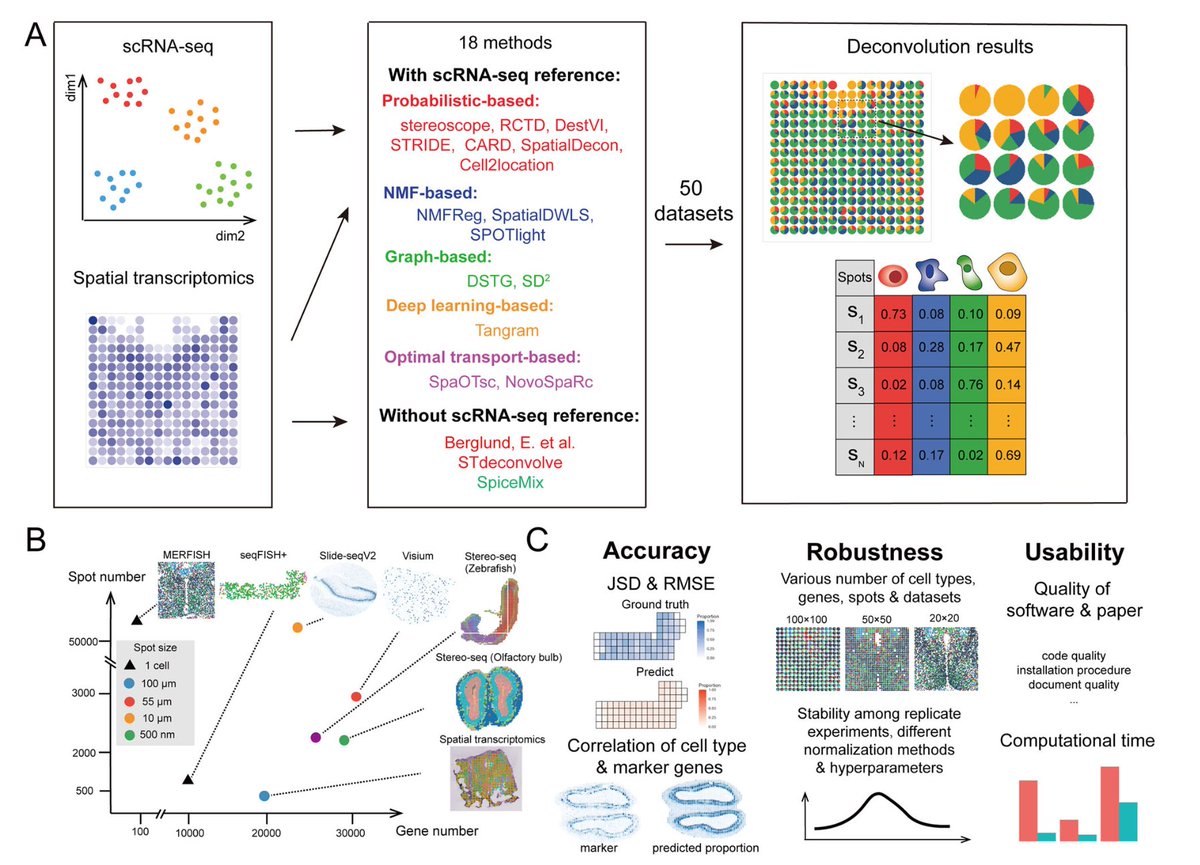
Do you need to analyze Spatial Transcriptomics data, but are lost in the endless sea of methods? Here's an explainer of the new @NatureComms paper benchmarking 18 spatial cellular deconvolution methods🧵🧵 nature.com/articles/s4146…
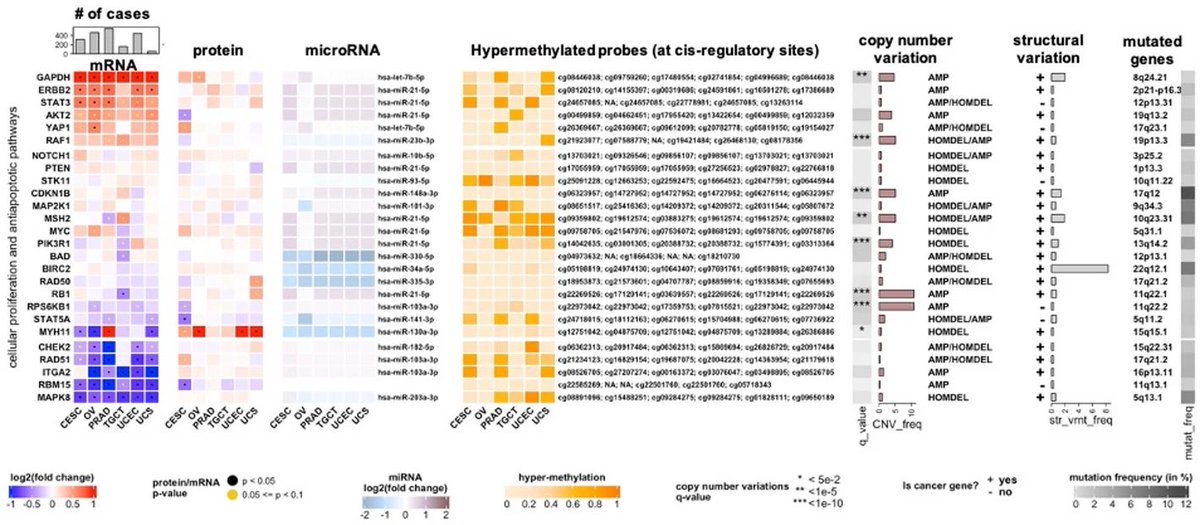
Impressive advancement in Computational Pathology. A new multimodal foundation model by @AI4Pathology trained on 47,000 paired histology & genomics, which beautifully shows the multi-modal power of images & DNA & RNA Even though patient genomic data is rare, it's so powerful 🧵

By pretraining on SpatialCorpus-110M, a curated resource of vast and diverse transcriptomes of dissociated and spatially resolved cells from both human and mouse, Nicheformer advances building foundation models for spatial single-cell analysis. nature.com/articles/s4159…

Our new method generates high-quality 3D multispectral point clouds for plants, improving close-range phenotyping. It enhances image fusion and corrects illumination, boosting accuracy. #PlantPhenotyping #SpectralImaging Details: doi.org/10.34133/plant…

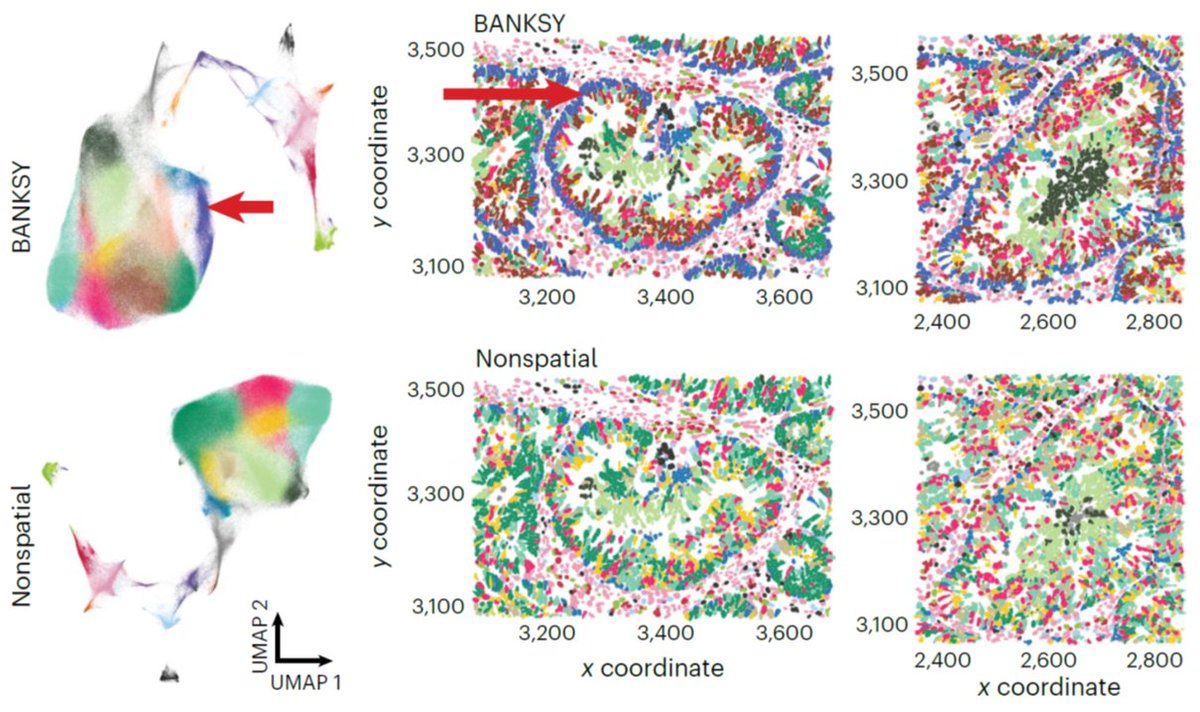
1. BANKSY is out! go.nature.com/4bUR200 Our spatial clustering algo applies to any spatial RNA/protein/... assay, scales to 2M cells, detects both cell typing and spatial domains and facilitates spatial batch correction. 🧵 👇
Systematic comparison of 11 sequencing-based #SpatialTranscriptomics methods A must-read for spatial omics researchers🤠 Visium DynaSpatial HDST BMKMANU S1000 Slide-seq V2 Curio Seeker Slide-tag Stereo-seq PIXEL-seq Salus DBiT-seq Guess which behaves best…

I'm so excited to finally introduce Fields of The World (FTW!) - the largest & most diverse ML dataset for field instance segmentation in satellite images. FTW has >70k samples from 24 countries (4 continents), with semantic & instance labels + Sentinel-2 RGB-NIR from 2 dates. 🧵

Something went wrong.
Something went wrong.
United States Trends
- 1. Marshawn Kneeland 21.5K posts
- 2. Nancy Pelosi 25.4K posts
- 3. #MichaelMovie 35.7K posts
- 4. #NO1ShinesLikeHongjoong 26.9K posts
- 5. #영원한_넘버원캡틴쭝_생일 26.6K posts
- 6. ESPN Bet 2,319 posts
- 7. Gremlins 3 2,957 posts
- 8. #thursdayvibes 2,969 posts
- 9. Madam Speaker 1,061 posts
- 10. Baxcalibur 3,680 posts
- 11. Chimecho 5,384 posts
- 12. Joe Dante N/A
- 13. Jaafar 10.8K posts
- 14. Chris Columbus 2,615 posts
- 15. Good Thursday 36.2K posts
- 16. #LosdeSiemprePorelNO N/A
- 17. Votar No 26.9K posts
- 18. Penn 9,260 posts
- 19. Diantha 1,537 posts
- 20. Korrina 4,241 posts