#xcms search results
X-Vision’25 ignited the brightest first-year minds from the Faculty of Commerce & Management, SXUK! Organized by XCMS, the event featured three exciting rounds — Prism Breaker (PR), Assets Aurora (Finance), & Spectrum Cipher (HR). #XVision25 #XCMS #SXUK
Twenty years in the making, the #XCMS-#METLIN integrated platform is now available with the complete METLIN MS/MS 935K+ molecular standards database. Available as a local version on your own personal computer. For information visit massconsortium.com
#ItsFreeMetabolomics But then again, what is... @mzmine_project, @GNPS_UCSD, @OpenMSTeam, #xcms, SIRIUS (@boeckerlab ), #massbank, ... Please post your favorite open source/free tools.

Robust growth in #metabolomics and #lipidomics over the last 7 years as represented by the daily analyses performed on #XCMS Online. Thank you Paul Benton @scrippsresearch for providing the graphic.

As all three #LCMS are down 😰 I've had to seek productivity elsewhere: testing the fidelity of some #XCMS chemoinformatic analysis and having fun in the lab with dangerous chemicals. More method development for the stool analysis I'm working on. #GCMS

import #XCMS/CAMERA data (395x31100) + QC-SVRC + clean-up + import 22000 MSMS spectra & 241952 HMDB spectra + metabolite annotation of ~1500 features, in 5 lines of #MATLAB code and 10 min #freetime #metabolomics @j_kuligowski @IISLaFe
#Metabolomics enthusiasts, fasten your seat belts the 24th April (Tues) 11 am EST,@MetabolomicsSoc Webinar Prof. Oliver Fiehn @WCMC_UCDavis speak "Use better #software: MS-DIAL instead of #XCMS, ChemRich/MetDA instead of #Metaboanalyst " #Register here: bit.ly/2Hk8fpl

1st time attending #SETACSciCon!🎉Don't miss our poster on the occurrence and trend of emerging contaminants in Asopos river basin using #HRMS, #XCMS and #rstats💻 Join us for discussion💬@SETAC_world @reza_aalizadeh @MaristinaNika @nikos_thomaidis @ThomaidisLab

First time in my office today (since March) to pick up stuff - most importantly: #MSnbase and #xcms stickers 😅

Far far beyond #metabolomics, #massspec, and even #XCMS and #METLIN. Learning some new skills, just in case, and helping a friend put a roof on at a farm. My first experience with a pneumatic nailer and a 4:12 roof. Both enjoyable & the final product looked pretty good.

New beginnings @scrippsresearch (now w intended video) connecting main #metabolomics lab w our AOA (Advanced Open Access) MS lab. Populated w @ThermoFisher, @Bruker, @Agilent & @Waters, high-end MS experience for Scripps students & post docs, of course including #XCMS #METLIN
New beginnings @scrippsresearch, just connected our main #metabolomics MS lab with the AOA (Advanced Open Access) lab (except for the plastic). Populated w @ThermoFisher, @Bruker, @Agilent & @WatersCorp high end MS for students and post docs. And, of course, #XCMS #METLIN

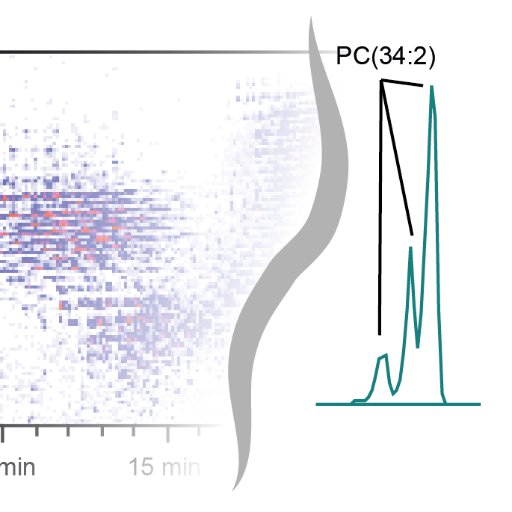
One of many lipids observed in a fasting study. Too early to know its significance, but I can't complain about the aesthetics of the EIC and the consistency between the different level of fasted groups. Well done Winnie, expanding into applying #XCMS & #METLIN

Winnie has run over a million molecular standards for the #METLIN MS/MS database. So, I asked, how she maintained her sanity? Her response (using her preferred “hand drawn” filter), in short, vacations and the satisfaction of creating high quality data.
Data processing, #multi-omic pathway mapping, and #metabolite activity analysis using #XCMS Online: go.nature.com/2HU3Qpy from @kadzuis @scrippsresearch #visualization #metabolomics #analytics #bigdata #multiomics #integratedomics




The simplistic beauty of #XCMS-#METLIN precursor ion filtering combined with MS/MS matching to deconvolve data on 935K+ molecular standards to a couple of hits @MolSystBiol doi.org/10.1038/s44320… XCMS-METLIN local for your PC now available at massconsortium.com
Twenty years in the making, the #XCMS-#METLIN integrated platform is now available with the complete METLIN MS/MS 935K+ molecular standards database. Available as a local version on your own personal computer. For information visit massconsortium.com
@jo_rainer kicks off the #SWEMSA #metabolomics session chaired by @wittingmichael1 by highlighting recent updates in #XCMS!


New beginnings @scrippsresearch, just connected our main #metabolomics MS lab with the AOA (Advanced Open Access) lab (except for the plastic). Populated w @ThermoFisher, @Bruker, @Agilent & @WatersCorp high end MS for students and post docs. And, of course, #XCMS #METLIN

ppm, snthresh, mzdiff, prefilter, etc - Peak picking parametrisation can be challenging! Check out this new application for LC-MS #data_viz and parameter fine-tuning of #xcms peak picking algorithms #msbrowser #metabolomics #rstats tkimhofer.github.io/msbrowser/
X-Vision’25 ignited the brightest first-year minds from the Faculty of Commerce & Management, SXUK! Organized by XCMS, the event featured three exciting rounds — Prism Breaker (PR), Assets Aurora (Finance), & Spectrum Cipher (HR). #XVision25 #XCMS #SXUK
A free version just posted—courtesy of @Nature and @JaviGBermudez on cancer lipidomics. rdcu.be/eqx3e Lipidomics (via #XCMS/#METLIN) reveals α-tocopherol (vitamin E) as a key player in mediating cancer cell death. #Lipidomics #CancerResearch #Ferroptosis #VitaminE
Lipidomics (& #METLIN) reveal a metabolic vulnerability in tumors @Nature doi.org/10.1038/s41586… Tumors don’t just synthesize their own lipids—they scavenge antioxidant-rich lipoproteins. @JaviGBermudez w/ our lab's lipidomics uncover how this can be used to impair tumor growth
The simplistic beauty of #XCMS-#METLIN precursor ion filtering combined with MS/MS matching to deconvolve data on 935K+ molecular standards to a couple of hits @MolSystBiol doi.org/10.1038/s44320… XCMS-METLIN local for your PC now available at massconsortium.com
Twenty years in the making, the #XCMS-#METLIN integrated platform is now available with the complete METLIN MS/MS 935K+ molecular standards database. Available as a local version on your own personal computer. For information visit massconsortium.com
Twenty years in the making, the #XCMS-#METLIN integrated platform is now available with the complete METLIN MS/MS 935K+ molecular standards database. Available as a local version on your own personal computer. For information visit massconsortium.com
Sexual communication signals identified by @AndiFischer10 using untargeted #ActivityMetabolomics @SciReports nature.com/articles/s4158… nature.com/articles/s4159… #XCMS @AndiFischer10 narrows down the active pheromones in spiders. Nice work Andi!
nature.com
Non-targeted metabolomics aids in sex pheromone identification: a proof-of-concept study with the...
Scientific Reports - Non-targeted metabolomics aids in sex pheromone identification: a proof-of-concept study with the triangulate cobweb spider, Steatoda triangulosa
One of many lipids observed in a fasting study. Too early to know its significance, but I can't complain about the aesthetics of the EIC and the consistency between the different level of fasted groups. Well done Winnie, expanding into applying #XCMS & #METLIN

Winnie has run over a million molecular standards for the #METLIN MS/MS database. So, I asked, how she maintained her sanity? Her response (using her preferred “hand drawn” filter), in short, vacations and the satisfaction of creating high quality data.
Far far beyond #metabolomics, #massspec, and even #XCMS and #METLIN. Learning some new skills, just in case, and helping a friend put a roof on at a farm. My first experience with a pneumatic nailer and a 4:12 roof. Both enjoyable & the final product looked pretty good.

#0day #XCMS v1.83 - Remote Command Execution #Exploit #RCE 0day.today/exploit/descri…
My recent PR to the #xcms @Bioconductor package has ID 666 👹 no surprise: it comes with some devilishly new functionality: - fully based on #Spectra package - under the hood changes to increase performance for some steps - ... github.com/sneumann/xcms/…
github.com
xcms 4: native support for MsExperiment and Spectra by jorainer · Pull Request #666 · sneumann/xcms
This PR adds native support for Spectra to xcms: in addition to a OnDiskMSnExp it is now possible to run the full xcms pre-processing on a MsExperiment object that bases/uses Spectra for MS data re...
New beginnings @scrippsresearch (now w intended video) connecting main #metabolomics lab w our AOA (Advanced Open Access) MS lab. Populated w @ThermoFisher, @Bruker, @Agilent & @Waters, high-end MS experience for Scripps students & post docs, of course including #XCMS #METLIN
New beginnings @scrippsresearch, just connected our main #metabolomics MS lab with the AOA (Advanced Open Access) lab (except for the plastic). Populated w @ThermoFisher, @Bruker, @Agilent & @WatersCorp high end MS for students and post docs. And, of course, #XCMS #METLIN

New beginnings @scrippsresearch, just connected our main #metabolomics MS lab with the AOA (Advanced Open Access) lab (except for the plastic). Populated w @ThermoFisher, @Bruker, @Agilent & @WatersCorp high end MS for students and post docs. And, of course, #XCMS #METLIN

One of the architects of #RforMassSpectrometry @rstatstweet #xcms #MSnbase #Spectra @lgatt0 @sneumannoffice
We stay in the drylab with @jo_rainer from @EURAC! He has an open position in #computational #metabolomics focusing on methods facilitating handling, processing and annotation of #metabolomics data

I didn’t think @lizwant solvent story @an_chem on #XCMS #METLIN deconvolving extraction/distribution would get much attention. However due to her attention to detail & excellent science it still sees a lot of interest, nice work Prof Liz! @imperialcollege doi.org/10.1021/ac0513…
pubs.acs.org
Solvent-Dependent Metabolite Distribution, Clustering, and Protein Extraction for Serum Profiling...
The aim of metabolite profiling is to monitor all metabolites within a biological sample for applications in basic biochemical research as well as pharmacokinetic studies and biomarker discovery....
Though question. Don't want to offend anyone, but aside from new MS instrumentation/tech, I think the most succesful/accepted advances were new developments based on #mzmine and #xcms plus all the growing MS/MS databases. Yet, many others planted the seeds for 2020-2025 advances.
Thank you @longlabstanford & great to hear that #XCMS & #METLIN could help in this excellent @Nature doi.org/10.1038/s41586… research. Here are two sources for the Lac-Phe for those interested (as I'm sure there will be many) trc-canada.com/product-detail… novoprolabs.com/p/lac-phe-3189…
Metlin was really key for us during the process of structure ID!!!
Noteworthy updates in the #xcms package part of @Bioconductor release 3.15: - m/z of ROI is calculated using intensity weighted mean (see github.com/sneumann/xcms/… for details). Thanks to @JanStanstrup for reporting. - functions for subsetting rewritten.
github.com
Fix peak mz by jorainer · Pull Request #606 · sneumann/xcms
This PR changes the way centWave calculates the m/z of a peak if within the ROI there are more than one mass peak per spectrum. In theory, for well centroided data, this should not happen but can h...
Paramounter currently works for #XCMS, #MSDIAL, and #MZmine2-based data processing. Support for #OpenMS and #MAVEN is in the works.
Correlated Ion Monitoring is a personal favorite w @MartinGiera & Rico Derks. Rico borrowed from #XCMS to empower single quads to do triple quad MRM, or Q-MRM, this time for peptides pubs.acs.org/doi/10.1021/ja…. Talent runs deep at @LUMC_Leiden
pubs.acs.org
Proteomics with Enhanced In-Source Fragmentation/Annotation: Applying XCMS-EISA Informatics and...
Enhanced in-source fragmentation/annotation (EISA) has recently been shown to produce fragment ions that match tandem mass spectrometry data across a wide range of small molecules. EISA has been...
True @JustBillings, yet the team hasn't been sleeping, multiple #METLIN #XCMS @JeanGalano developments coming related to @CaymanChemical, ion mobility, neutral loss, machine learning and similarity analysis. And my (& @MartinGiera's) personal favorite Q-MRM.
#ItsFreeMetabolomics But then again, what is... @mzmine_project, @GNPS_UCSD, @OpenMSTeam, #xcms, SIRIUS (@boeckerlab ), #massbank, ... Please post your favorite open source/free tools.

Robust growth in #metabolomics and #lipidomics over the last 7 years as represented by the daily analyses performed on #XCMS Online. Thank you Paul Benton @scrippsresearch for providing the graphic.

import #XCMS/CAMERA data (395x31100) + QC-SVRC + clean-up + import 22000 MSMS spectra & 241952 HMDB spectra + metabolite annotation of ~1500 features, in 5 lines of #MATLAB code and 10 min #freetime #metabolomics @j_kuligowski @IISLaFe
As all three #LCMS are down 😰 I've had to seek productivity elsewhere: testing the fidelity of some #XCMS chemoinformatic analysis and having fun in the lab with dangerous chemicals. More method development for the stool analysis I'm working on. #GCMS

Using #XCMS, #METLIN, and #LipidMAPS to identify unknown metabolites in untargeted LC-MS #UABMetabolomics2017




First time in my office today (since March) to pick up stuff - most importantly: #MSnbase and #xcms stickers 😅

#Metabolomics enthusiasts, fasten your seat belts the 24th April (Tues) 11 am EST,@MetabolomicsSoc Webinar Prof. Oliver Fiehn @WCMC_UCDavis speak "Use better #software: MS-DIAL instead of #XCMS, ChemRich/MetDA instead of #Metaboanalyst " #Register here: bit.ly/2Hk8fpl

@jo_rainer kicks off the #SWEMSA #metabolomics session chaired by @wittingmichael1 by highlighting recent updates in #XCMS!


Data processing, #multi-omic pathway mapping, and #metabolite activity analysis using #XCMS Online: go.nature.com/2HU3Qpy from @kadzuis @scrippsresearch #visualization #metabolomics #analytics #bigdata #multiomics #integratedomics




Paul Benton and I introducing metabolomics and XCMS at the Proteomics Society Symposium workshop #metabolomics #mexico #XCMS

Something went wrong.
Something went wrong.
United States Trends
- 1. Marshawn Kneeland 44K posts
- 2. Nancy Pelosi 64.6K posts
- 3. Craig Stammen 1,772 posts
- 4. Gordon Findlay 2,259 posts
- 5. Ozempic 5,780 posts
- 6. Michael Jackson 68.5K posts
- 7. Pujols N/A
- 8. #ThankYouNancy 1,478 posts
- 9. Novo Nordisk 5,971 posts
- 10. GLP-1 4,670 posts
- 11. Abraham Accords 4,279 posts
- 12. #NO1ShinesLikeHongjoong 37.4K posts
- 13. Kazakhstan 6,019 posts
- 14. Kyrou N/A
- 15. #영원한_넘버원캡틴쭝_생일 36.6K posts
- 16. Preller N/A
- 17. Gremlins 3 4,898 posts
- 18. Kinley N/A
- 19. Unplanned 9,115 posts
- 20. Joe Dante N/A