Yang Lee
@leetle_yang
Lab boffin de l'ordinaire @NxeraPharma formerly @MRC_LMB | Taking tips from tip boxes in the (only) correct order | #CryoEM #GPCR
Was dir gefallen könnte
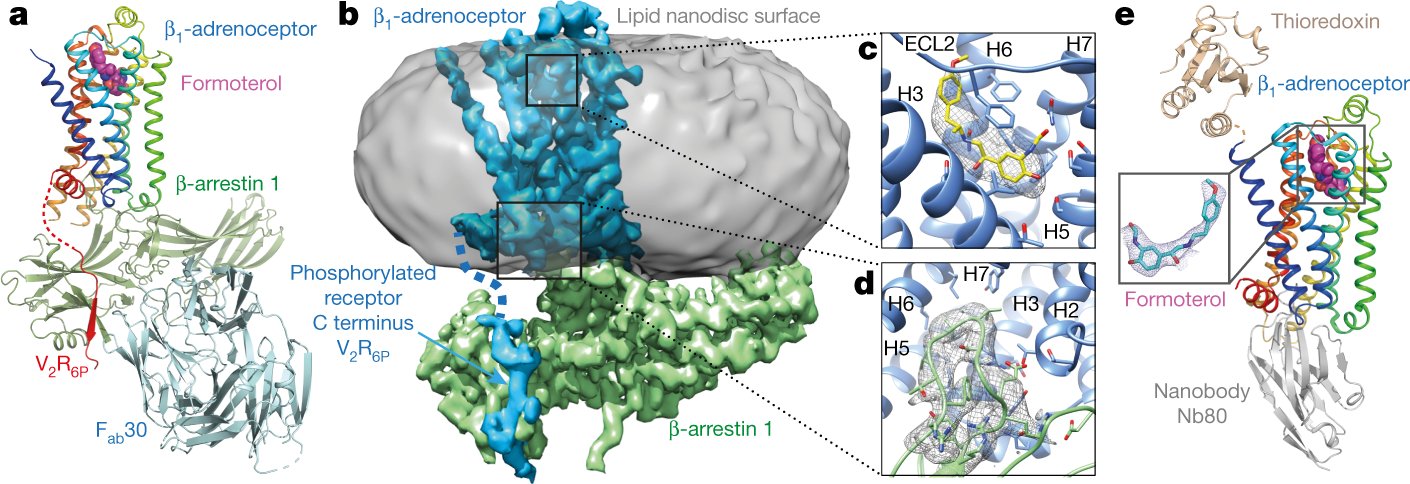
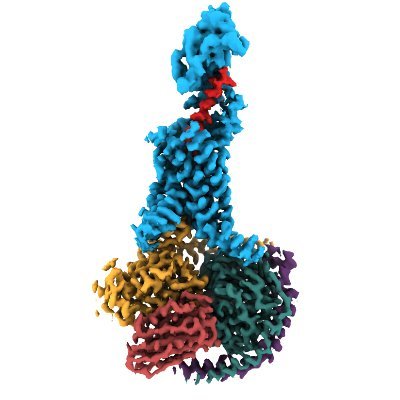
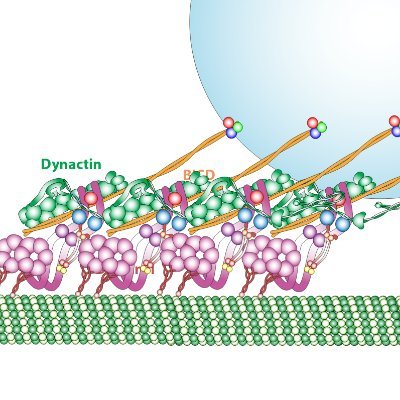
The Tate group @MRC_LMB is happy to contribute to our collective understanding of arrestin-coupling to #GPCRs. We present a #cryoEM model of the β1AR-βarr1 complex bound to formoterol, an arrestin-biased agonist, in nanodisc. @JGarciaNafria @arshukla nature.com/articles/s4158…
Excited to share our AI+cryo-EM work! 🧬 🔬 Cryo-IEF: Foundation model trained on 65M particles 🤖 CryoWizard: automated structure pipeline 🎯 Making cryo-EM accessible to more labs Preprint: biorxiv.org/content/10.110… Code: github.com/westlake-repl/… #CryoEM #AI #StructuralBiology


Now online: We created de novo insulin receptor agonists using computational protein design! @EunheeChoi12 @XiaochenB @PreethamVi @UWproteindesign #BakerLab biorxiv.org/content/10.110…

De novo Design of A Fusion Protein Tool for GPCR Research biorxiv.org/cgi/content/sh… #biorxiv_biophys
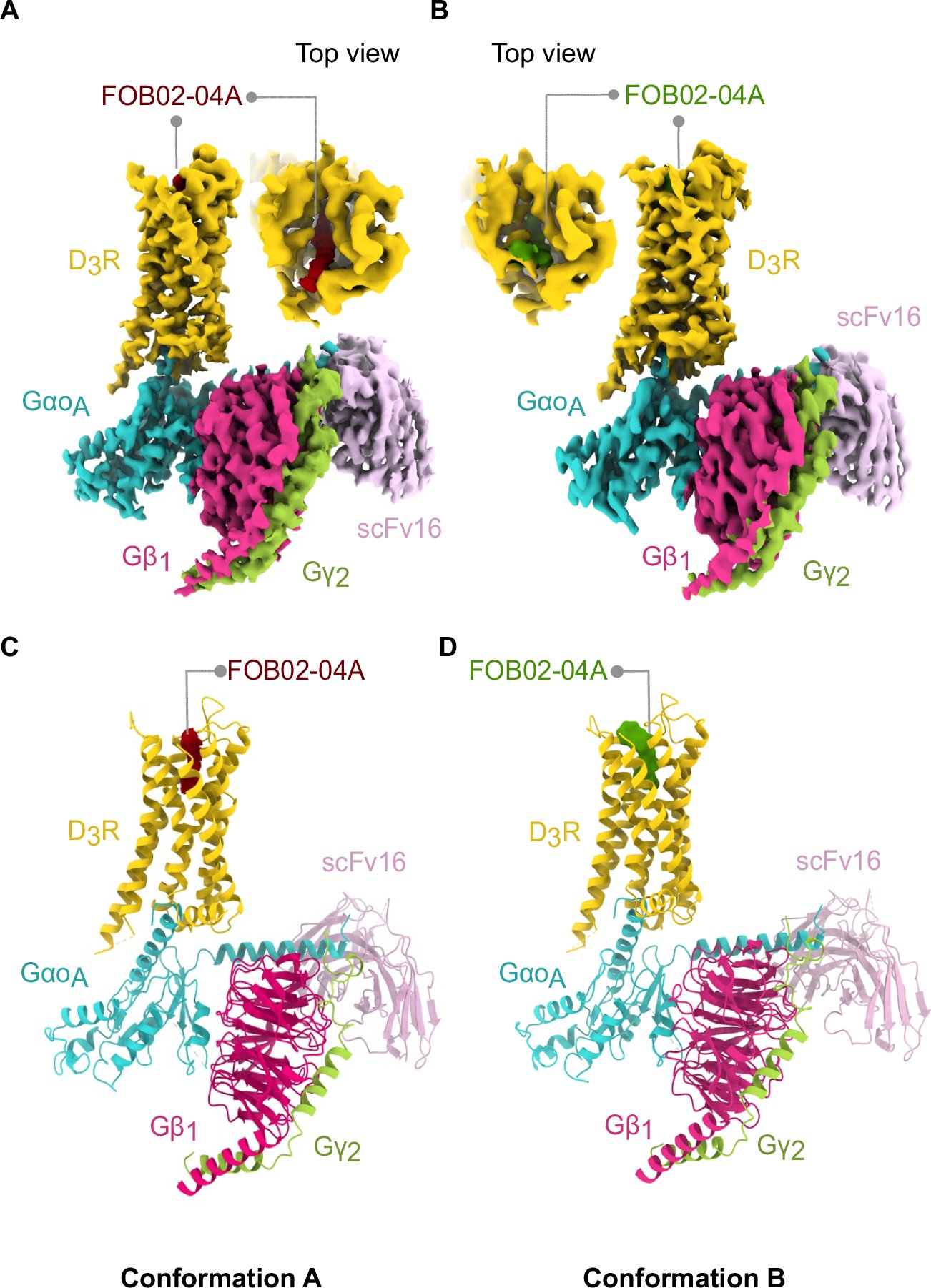
Delighted to get this out!we describe bitopic binding mode to the dopamine 3 receptor, including a reorganization of TM1 that generates a unique site that could be exploited to develop selective molecules. Tremendous effort from @sandrarrour @angela31398 nature.com/articles/s4146…
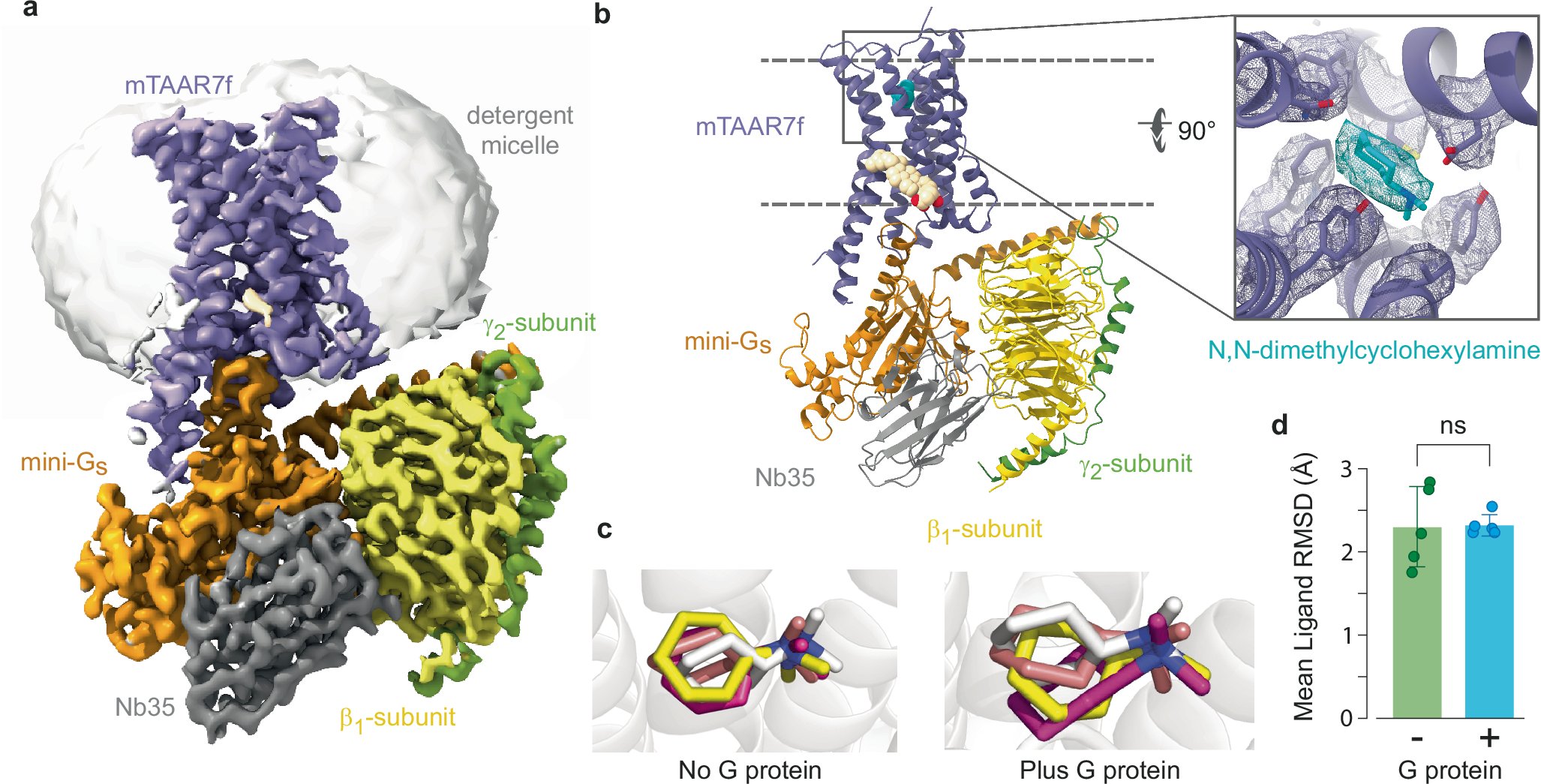
How to sense danger from afar? Revised and refined, our paper on the insights into smell recognition by a mammalian trace-amine receptor from Cryo-EM, modelling and functional studies is finally out! nature.com/articles/s4146…
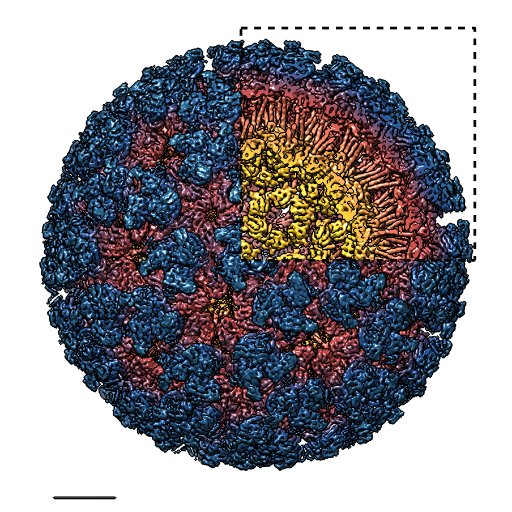
#cryoEM with energy filter & electron counting significantly increased resolution even for a full size protein like proteinase K. Sub-atomic resolution #MicroED rivals best reported Xray of Proteinase K but by using crystals a billionth the size. A section of the map is shown

Cryo-EM phase-plate images reveal unexpected levels of apparent specimen damage biorxiv.org/cgi/content/sh… #biorxiv_biophys
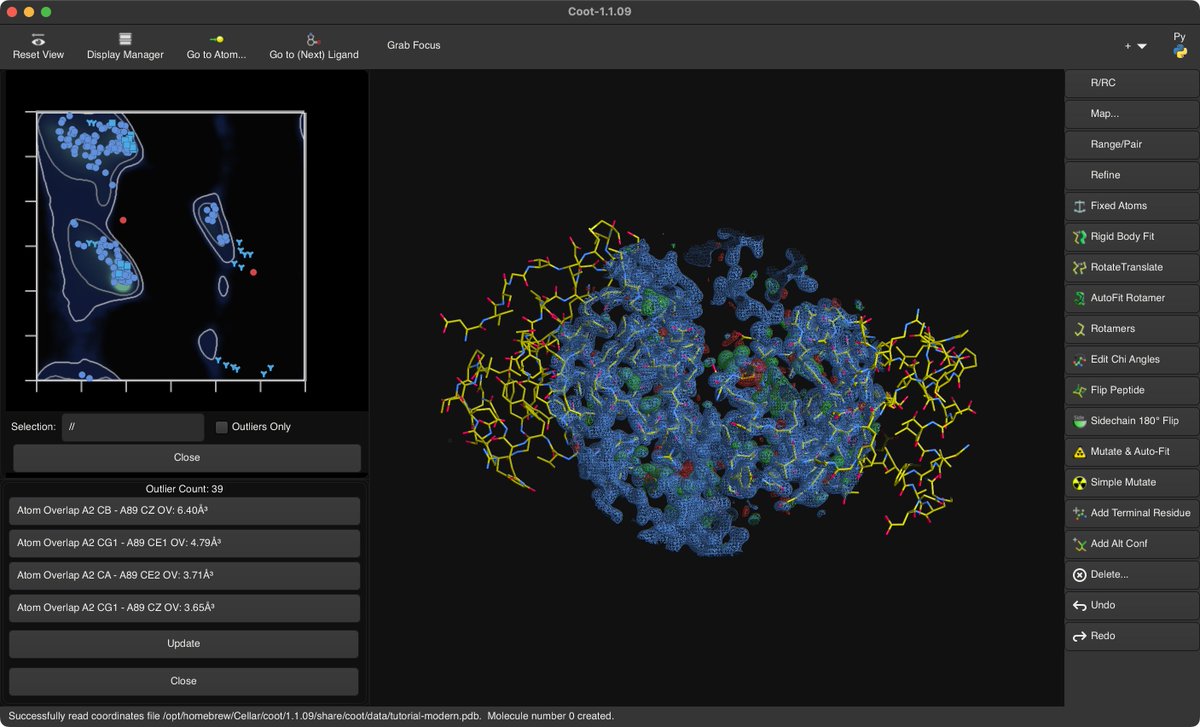
🎉 New formula Coot 1.1.09 in Brewsci/bio for Linux and macOS! ℹ️ Crystallographic Object-Oriented Toolkit 🍺 brew install brewsci/bio/coot #Bioinformatics
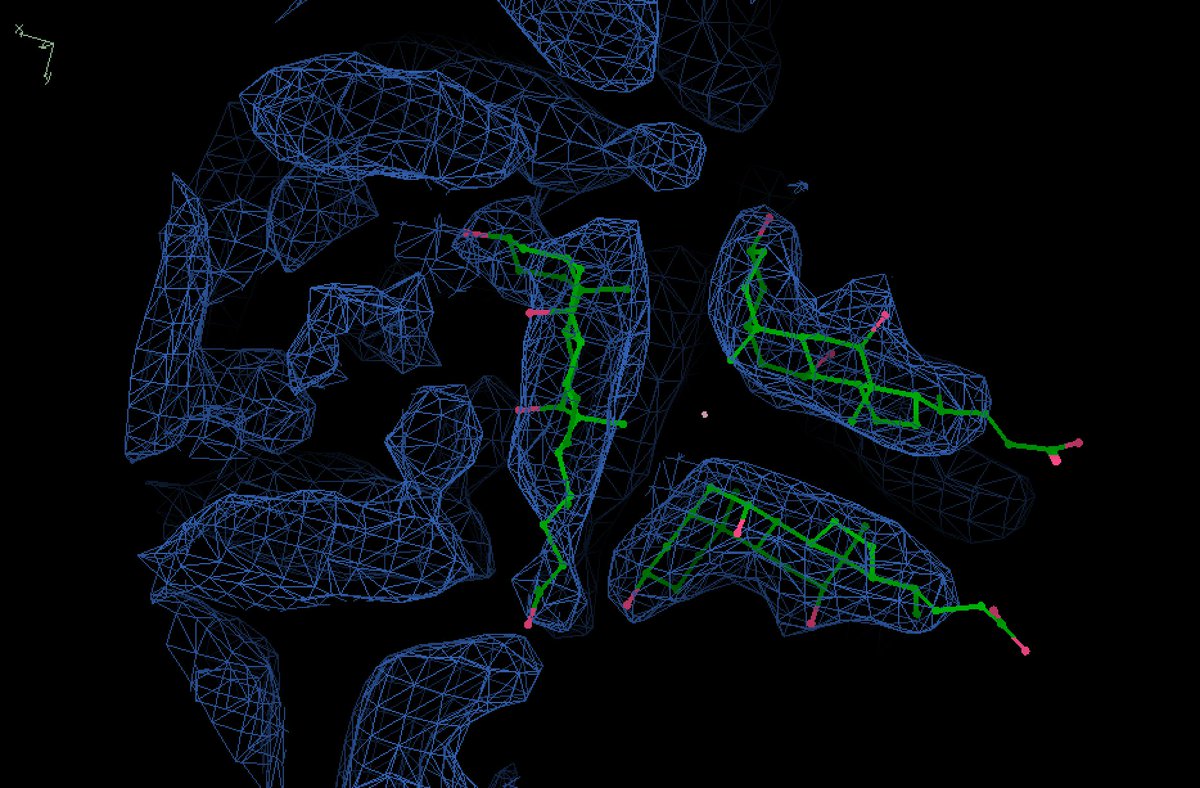
Final model for this weird detergent filament, with a helix of H-bonded water molecules at the center:
Ok it is indeed a helical filament of deoxycholate:
Obviously Deep Mind/Isomorphic have commercial interests… but they need not seek the faux legitimacy of a Nature paper absent the transparency essential for vetting. And Nature wants to abandon their policies to game their impact factor and steal cheap attention with what will…
The latest version of AlphaFold models how proteins interact with other molecules — but DeepMind restricts access to the tool go.nature.com/4dsMcYP
Although it is a very exciting development, it is sad to see that no code will be released, which would not be allowed for academic institutions @nature. It was clear that weights will not be released (despite being trained purely on public data) but this I did not expect.
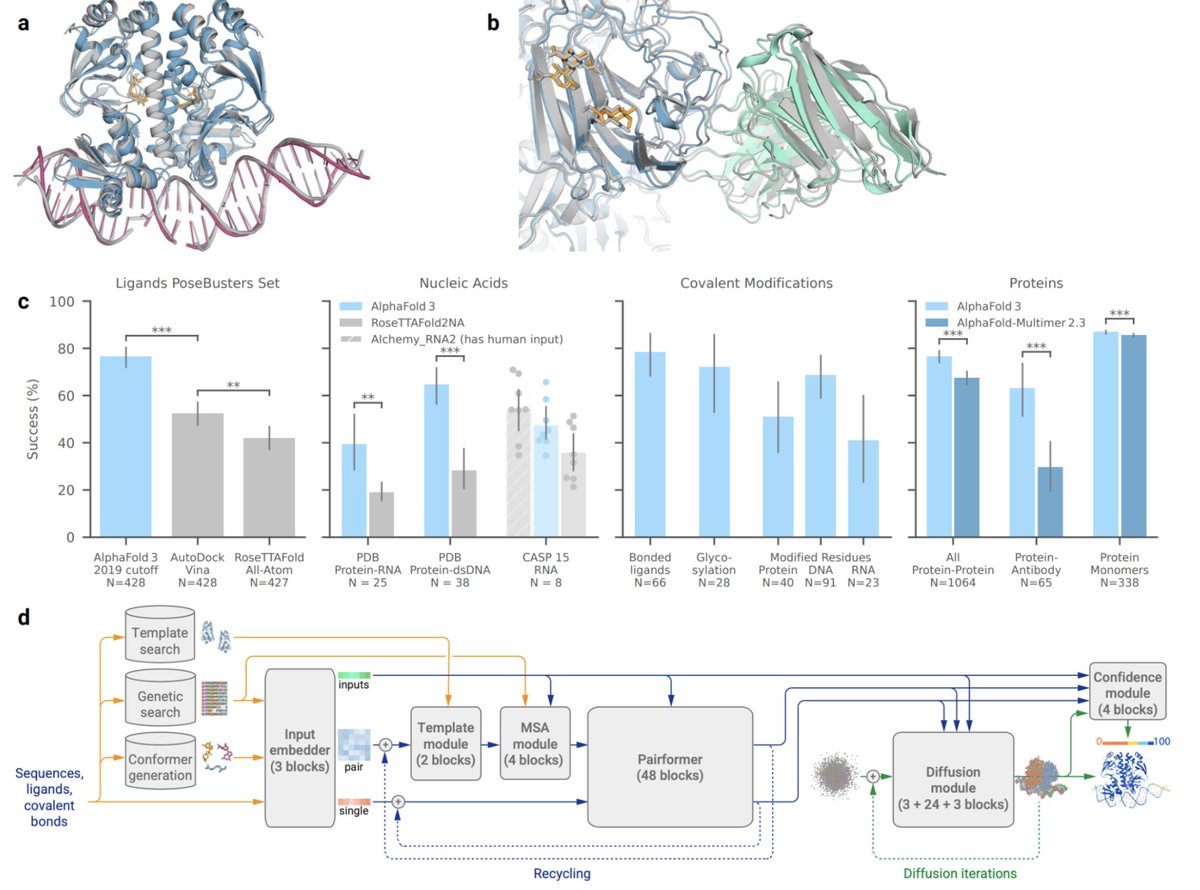
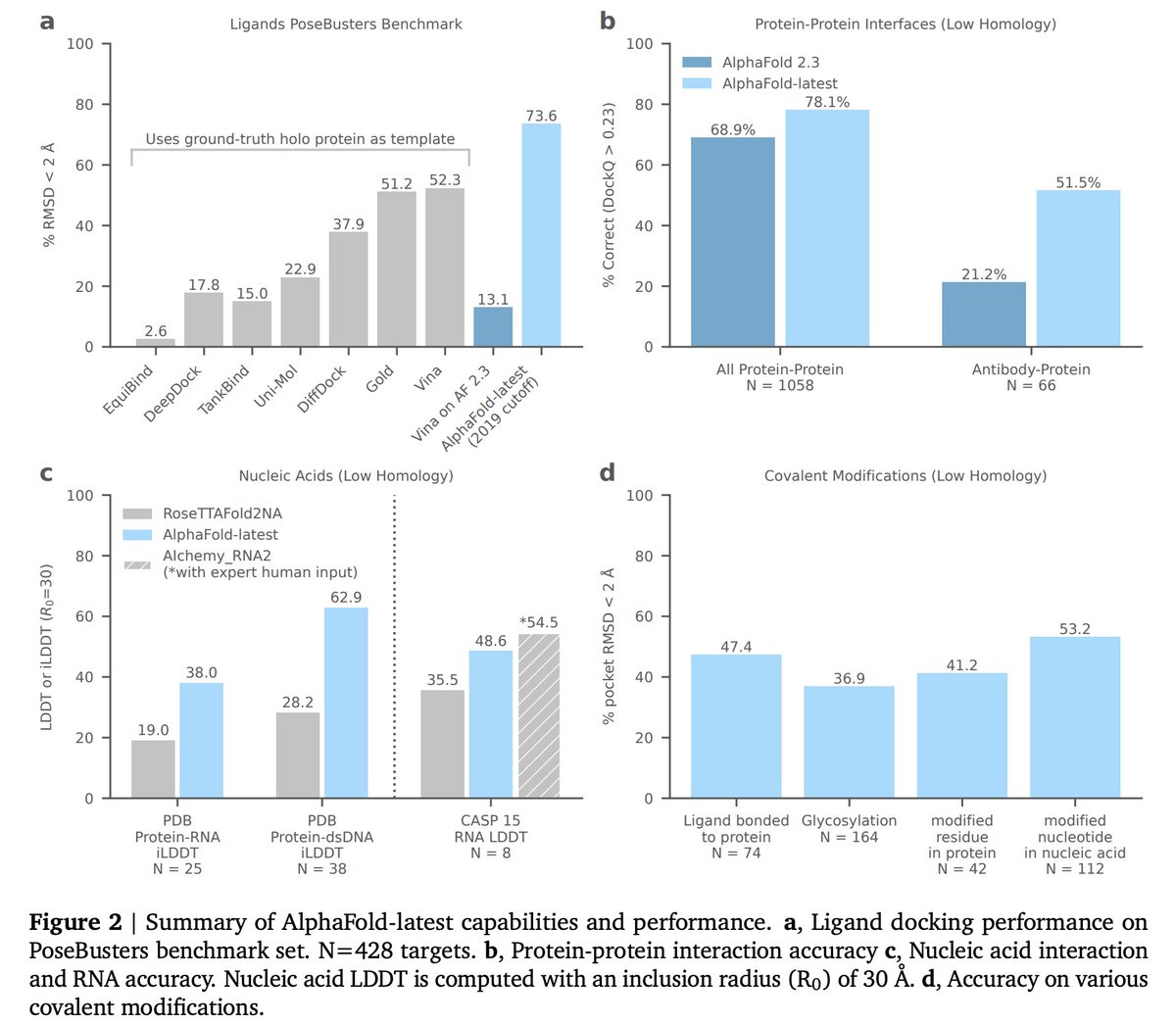
Super excited to be releasing AlphaFold 3 today, developed by @IsomorphicLabs and @GoogleDeepMind: our next generation AI model for predicting the biomolecular structures and interactions of proteins, DNA, RNA, small molecules, and more: bit.ly/44yfaCw 1/
Great work, beautiful structures, and very nice findings, congrats! I also invite everybody to check how this orthosteric invasion of a GPCR by cholesterol was already described some years ago, and suggested being feasible in other GPCRs 😉 nature.com/articles/ncomm…
Excited to read this from @yuntaoinnit in the @zhou_lab - saw Hong present it at last year's 3DEM GRC and have been waiting to try it out, look forward to trying spIsoNet on our own data soon! 🤩 biorxiv.org/content/10.110…

Excited to share our latest pre-print! We map GPCR activation probability at the plasma membrane of live cells. Strikingly, we see ultra-long-lived (~5 min) nanodomains of high activation probability, while most receptors remain inactive. Interested? 🧵shorturl.at/fkoBV
What about the issue with multiple glow discharge induced damage on recycled grids ?
#SerialEM 4.1 released as stable tonight, highlights include autocontouring of grid squares, hole finding on gold grids with a hex array of holes, saving of shifts from Shift to Marker, 118 new script commands, ... thank you for the tireless work, #stdavidmastronarde! 🤩

The latest model can predict 3D structures for complexes that include proteins, ligands, nucleic acids, ions, and modified residues. We report results across molecule-type benchmarks showing SOTA performance for protein-ligand, protein-RNA/DNA, protein-protein interfaces 2/n
Our EMMIVox approach for accurate model and ensemble refinement using #cryoEM maps and Bayesian inference is out in @biorxivpreprint Work led by Samuel Hoff @institutpasteur @DBSC_IP with contributions from @EmilThomasen and @LindorffLarsen biorxiv.org/content/10.110…
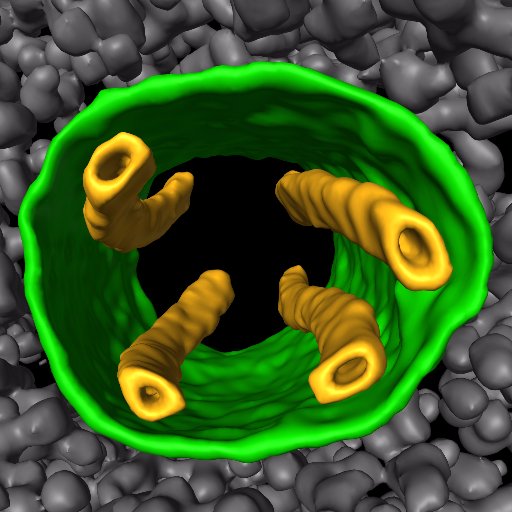
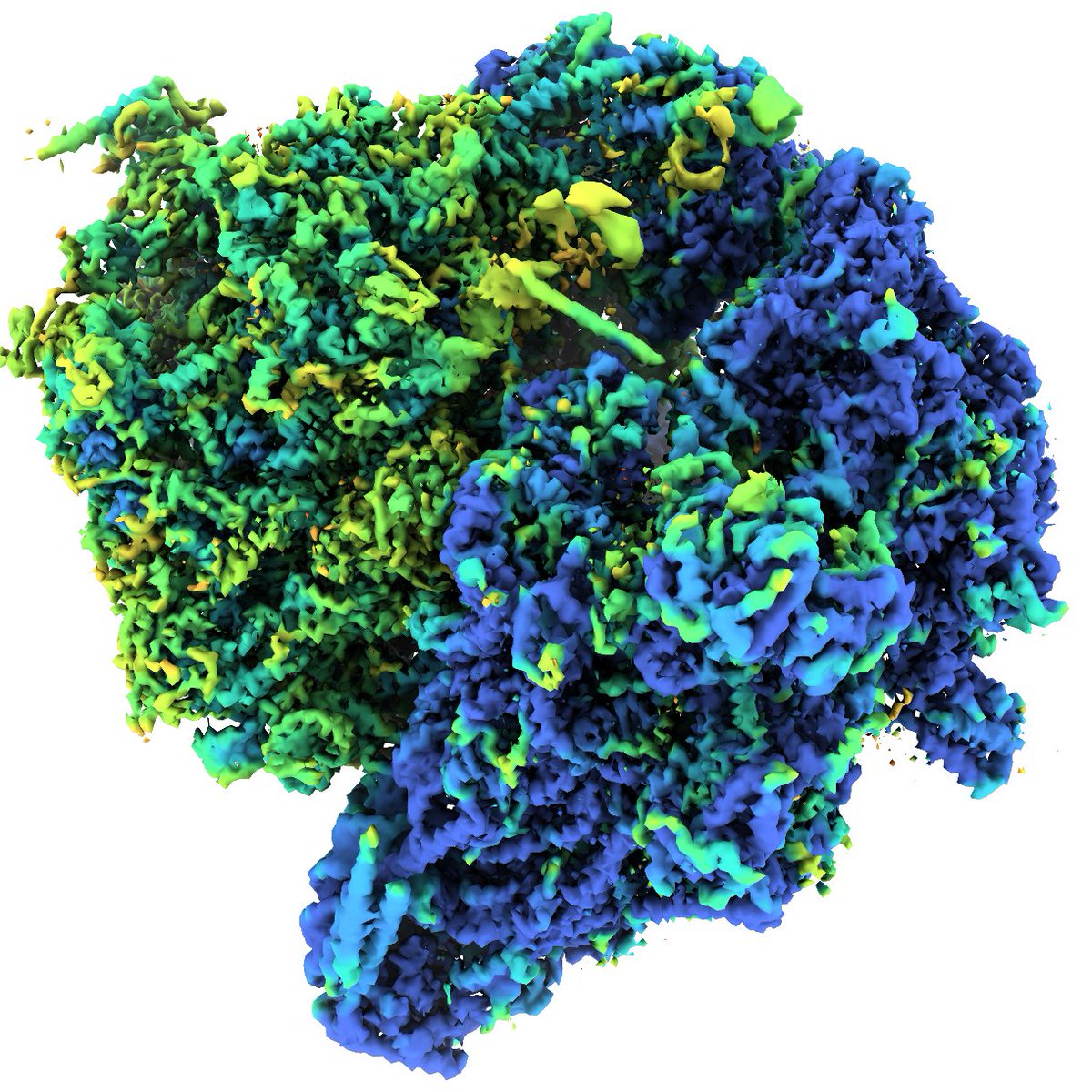
Me, @proteincapsid and @AlisterBurt made a tool to estimate composition in cryo-EM maps. Here for instance, one ribosome subunit is clearly lower occupancy. It's not a complicated tool, but then again it's only the start.
United States Trends
- 1. Zeraora 7,739 posts
- 2. Peggy 26.5K posts
- 3. #FaithFreedomNigeria 1,369 posts
- 4. Berseria 2,108 posts
- 5. Good Wednesday 33.1K posts
- 6. Luxray 1,298 posts
- 7. #wednesdaymotivation 7,013 posts
- 8. Dearborn 326K posts
- 9. #Wednesdayvibe 2,346 posts
- 10. Hump Day 16.2K posts
- 11. #LosVolvieronAEngañar 1,827 posts
- 12. Cory Mills 18.2K posts
- 13. #MissUniverse 23.9K posts
- 14. Happy Hump 10.5K posts
- 15. Jessica Tisch N/A
- 16. $NVDA 40.6K posts
- 17. Tom Steyer N/A
- 18. Semrush N/A
- 19. Ternopil 42.5K posts
- 20. For God 222K posts
Was dir gefallen könnte
Something went wrong.
Something went wrong.