#transcriptome نتائج البحث
BrainSTEM Atlas Maps Every Cell of the Developing Human Brain Researchers have created one of the most detailed cellular maps of the developing human brain, analyzing nearly 680,000 cells at single-cell resolution. The BrainSTEM atlas traces how each cell type grows,…

"Only ~0.02%-3.1% of [a cell's] genome" is being transcribed at any given moment. Other interesting takeaways from this new paper: > If you pool together a bunch of cells of the SAME type (like primary immune cells from a mouse's spleen), and you measure the transcription for…
![NikoMcCarty's tweet image. "Only ~0.02%-3.1% of [a cell's] genome" is being transcribed at any given moment.
Other interesting takeaways from this new paper:
> If you pool together a bunch of cells of the SAME type (like primary immune cells from a mouse's spleen), and you measure the transcription for…](https://pbs.twimg.com/media/G2CFMG2a4AAGWYL.png)
![NikoMcCarty's tweet image. "Only ~0.02%-3.1% of [a cell's] genome" is being transcribed at any given moment.
Other interesting takeaways from this new paper:
> If you pool together a bunch of cells of the SAME type (like primary immune cells from a mouse's spleen), and you measure the transcription for…](https://pbs.twimg.com/media/G2CFMS2bIAA07DE.png)
Here is the transcriptomics insight: the bulk RNA-seq (Fig 1), single-cell RNA-seq (Fig. 2), spatial transcriptomics (Fig. 3), and the original organ (Fig. 4). (Feel free to cite with image credit to Bo Xia)




Interested in #phylogenetics using whole #genome/#transcriptome/#proteome data? @C__Hahn and I are very happy to share phylociraptor with you: It facilitates explorative and reproducible phylogenomic analyses of thousands of genes and genomes. Preprint: tinyurl.com/yc4f7v6d
🚀 ¡Nuevo seminario en marcha! 📅 24/11 | 🕒 15:00 (CET) 🎙️ @pepeluisrr 🧬 Making the most of your transcriptomic landscapes: A computational workflow to quantify activity of context-specific biological functions at the single-cell level 📹 f.mtr.cool/iixdwjxcde

GTEx papers are out! 7k transcriptomes from 44 tissues & the broadest catalog of eQTLs to date. rdcu.be/wFqQ

Fabulous & fascinating talk as always by @LucyKornblith at #THOR2023 on the curious case of the platelet in trauma and what’s happening with the platelet #transcriptome - always a delight to learn from you, Lucy!

Wrapping up an inspiring few days at the EMBO Workshop: The Epitranscriptome. So many great talks, new ideas, and discussions about how RNA modifications shape gene expression. #EMBO #EMBL #Epitranscriptome #m6A #RNA

Reveal What Retroviral RTs Miss. Unlock deeper RNA-seq insights with uMRT, including: 🔹 Low-abundance transcripts 🔹 High GC content regions 🔹 Unique isoforms 🔹 Non-coding RNAs ...and more Enhance your RNA-seq today! Read more in the comments below... #RNASeq #RNA #BioTech

Amazing work headed by Sokolova team on #phage tail-tape fused #RNAP structural analysis. Glad ONT-cappable seq proved useful to analyze the #transcriptome landscape. #Researchpaper in @NatureComms : bit.ly/TailRNAP #thermophilic #virus #microbiology #crystallography

📢 We are looking for a motivated candidate for a fully funded #PhD to analyze the #transcriptome of polyploid cells from tomato fruit 🍅 at the single cell level #scRNAseq @INRAE_BFP @nathy_go_7 Info here 👇 jobs.inrae.fr/ot-22230

Excited to share our latest manuscript! How does an RNA get transported in neuronal cells? What sequences within the RNA govern this? We used a massively parallel reporter assay to identify sequences necessary and sufficient for RNA localization. 🧵biorxiv.org/content/10.110…

Starting to assemble the Reverse Transcriptase (RT) sculpture on a stand (left). Close ups of RT’s RnaseH domain on right. The stand represents the path to discovery (50 years ago) that RT converts RNA to DNA, a process that challenged a mainstream scientific concept of the time.


The epitranscriptome formed by the growing number of modifications occurring within mRNA transcripts.

The epitranscriptome formed by the growing number of modifications occurring within mRNA transcripts.

📰| #InstitutesPCUV 🧬🔬I2SysBio co-leads the largest comparative study on methods analyzing data obtained by long-read sequencing of the human #transcriptome. Read more ⬇️ news.pcuv.es/en/home-pcuv-i…

IMPRS retreat kicks off with a great talk by @torbenheick on #RNA turnover and #transcriptome quality control. #science #phdlife

🎃 Trick or transcript? Bacteria rewire their RNA world under stress & infection. tRNA mods as dual-function sensors, phage tRNA slashers, hibernating ribosomes & haunted translation hubs. 🧬🧪 Full November 1st issue → open.substack.com/pub/rnamodifup…

1/ We conducted one of the largest whole-#transcriptome studies, analyzing 300 #MDD patients. #TRD patients showed global transcriptomic changes involving immune, epigenetic, and non-coding RNA dysregulation.
showing how immune, epithelial, and metabolic networks are altered. These findings help clarify disease mechanisms and may support future therapeutic strategies. 🔗doi.org/10.1590/1678-7… #periodontitis #transcriptome #geneexpression #regulatorynetworks #bioinformatics
Explore : *** Overview of multi-omics data. Multi-omics data include #Genome, #Transcriptome, #Epigenome, #Proteome, #Exposome and #Microbiome. *** , Investigate Figure pmc.ncbi.nlm.nih.gov/articles/PMC87…, report overview @grok

#VegetableRes First transcriptome of sweet pitaya peel reveals genes for cuticle biosynthesis, key for water retention in the desert cactus fruit. @MaximumAcademic @PlantPhys #Cactus #Transcriptome Details: maxapress.com/article/doi/10…

We conducted one of the largest whole-#transcriptome studies, analyzing 300 #MDD patients. The findings suggest that #TRD represents a distinct molecular subtype characterized by unique #immune and #epigenetic signatures. Here the article 👇 sciencedirect.com/science/articl…
New in the September issue of #G3journal: @yanjunzan and the team reanalyzed #transcriptome data from thousands of #maize to uncover the genetics underlying maize complex traits, and built a multi-tissue regulatory atlas for functional validation. buff.ly/bQMa6Gs

A research team published their findings in Horticulture Research. The study investigated how non-photosynthetic grapevine cells adapt to glucose deprivation. #DNA methylation #Transcriptome Details: doi.org/10.1093/hr/uha…

New in #G3journal: @yanjunzan and the team reanalyzed #transcriptome data from thousands of #maize panels to uncover the genetics underlying maize complex traits, and built a multi-tissue regulatory atlas for functional validation and genomic improvements. buff.ly/lmrsmHC

🐟#HighlyCited Cardiac #Transcriptome and Histology of the Heart of the Male Chinese Mitten #Crab (Eriocheir sinensis) under High-Temperature Stress 👉mdpi.com/2410-3888/9/3/… 🦀#heatshockprotein #histology #immune

#ForestryRes #transcriptome #salt #ethylene Nitraria sibirica thrives under salt stress, showing higher growth and photosynthesis rates. Transcriptome analysis reveals upregulated genes involved in carbon fixation. 🧬 @MaximumAcademic @JournalNJFU Details: maxapress.com/article/doi/10…

ICAR–IASRI is organizing an Online Training on Transcriptomic Data Analysis during September 23–29, 2025. Develop applied expertise in bioinformatics tools. Apply: forms.gle/5p7Q2T1cGJka2r… more details, please see: iasri.res.in/API/Content/Up… #icar #Transcriptome #DataAnalysis

Molecular dissection of Xinong 511 spike rachis response to Fusarium head blight infection Xiaoying Yang, Maoru Xu ... Chunhuan Chen, Wanquan Ji link.springer.com/article/10.100… #FHB #transcriptome #DiseaseResistance #PlantImmunity
🌸 What controls #flower formation in Camellia oleifera? New study reveals GA dynamics + FT–LFY module drive floral initiation in tea-oil tree buds 🧪 #Transcriptome + #hormone profiling = insights into key flowering genes 🔗 brnw.ch/21wUI9v #FloralDevelopment

Using integrated #transcriptome and proteome analyses, scientists from @EmoryMedicine identified and validated eight novel non-classical #CDMarkers that help characterize #MesenchymalStemCells and potentially discriminate #iMSCs from other cell types. @stmcdr #OpenAccess:…

Greehey CCRI PI, Gail Tomlinson, MD, PhD, co-authored a recent paper in #CancerEpidemiologyBiomarkersPrevention (@AACR), "#Multiancestry #Transcriptome-Wide Association Study Identifies Candidate #Genes Associated with #Hepatoblastoma." tinyurl.com/mwb94ahp #ChildhoodCancer

🌺 How does Camellia azalea flower in summer? Xu et al. uncover the transcriptomic & hormonal shifts driving its floral development, integrating gene expression & phytohormone analysis. 📖 brnw.ch/21wUyVn #Camellia #PlantMolecularBiology #Transcriptome #Flowering

In a study published in Horticulture Research, researcher performed full-length transcriptome sequencing on chili pepper tissues at five growth stages. #plant developmental biology #transcriptome sequencing Details: doi.org/10.1093/hr/uha…

Interested in #phylogenetics using whole #genome/#transcriptome/#proteome data? @C__Hahn and I are very happy to share phylociraptor with you: It facilitates explorative and reproducible phylogenomic analyses of thousands of genes and genomes. Preprint: tinyurl.com/yc4f7v6d

Article by @Miriam_science et al @arbona_vicent @rosarivar @CebasPlant @ThePlantJournal @SEBiology @wileyplantsci Specific #ABA-independent #tomato #transcriptome reprogramming under #abiotic stress combination onlinelibrary.wiley.com/doi/10.1111/tp… #PlantSci @BigPurpleTomato @transcriptomes

Centrohelid #heliozoan #transcriptome: no flagellar genes (not surprising) MT-severing proteins and kinesin duplication (cool! probably a role in superfast contraction). Ikeda et al. 2022 10.1111/jeu.12955; picture Ikeda et al. 2020 10.4467/16890027AP.20.001.12157.

Amazing paper title for an amazing paper: "The swan #genome and #transcriptome, it is not all black and white" #biology #science #evolution #omics 🔗: genomebiology.biomedcentral.com/articles/10.11…

IMPRS retreat kicks off with a great talk by @torbenheick on #RNA turnover and #transcriptome quality control. #science #phdlife

📢#CallForPapers on Integrative Physiology and Translational-Omics of Exercise and Physical Activity! ow.ly/ZmzX50Px0Yl #transcriptome #microbiome #metabolome #proteome #epigenome #AcademicTwitter

Fabulous & fascinating talk as always by @LucyKornblith at #THOR2023 on the curious case of the platelet in trauma and what’s happening with the platelet #transcriptome - always a delight to learn from you, Lucy!

#Heterosis works—but how? Liu et al. tackle this question in new research combining #transcriptome, #proteome, #physiological, and heterosis analyses to study #salt response in super hybrid #rice. doi.org/10.1111/jipb.1… @wileyplantsci #PlantSci #JIPB #CropSci

Amazing work headed by Sokolova team on #phage tail-tape fused #RNAP structural analysis. Glad ONT-cappable seq proved useful to analyze the #transcriptome landscape. #Researchpaper in @NatureComms : bit.ly/TailRNAP #thermophilic #virus #microbiology #crystallography

Measuring natural selection on the #transcriptome 📖 nph.onlinelibrary.wiley.com/doi/10.1111/np… Viewpoint article by Stinchcombe and Kelly @JohnStinchcombe @WileyPlantScience #PlantScience

We developed LSTrAP-denovo, a pipeline for generating transcriptome atlases for species without a genome. All you have to do is to enter the tax ID of your species of interest... #transcriptome #assembly #CDS

Co-chair @LesleyRGolden @universityofky presenting their work on mid-life #APOE4 to #APOE2 ‘Switching’ alters the cerebral #transcriptome and decreases AD neuropathology #ISMNDNextGen

Today @AROMWM come and learn about the #transcriptome and #epigenome of the human #innerear sensory and non-sensory epithelia using @10xGenomics Multiome. Poster SA80 presented by @EmiliaLuca_PhD @Sunnybrook @UofT_OHNS @uoftmedicine @LMP_UofT #ARO2023

In the next #plenary our invited speaker @anaconesa will discuss the potential and challenges of using single-molecule long-read sequencing for #transcriptome analysis and #genome annotation🧬 Join us to learn more! bit.ly/3qClsB2 @Conesa_Lab @BioGenEurope @i2sysbio

📢 We are looking for a motivated candidate for a fully funded #PhD to analyze the #transcriptome of polyploid cells from tomato fruit 🍅 at the single cell level #scRNAseq @INRAE_BFP @nathy_go_7 Info here 👇 jobs.inrae.fr/ot-22230

New in @The_InnovationJ! A high-definition spatiotemporal transcriptomic atlas of mammalian kidney development. Jiang et al. present a spatially resolved #transcriptome atlas at single-cell resolution to explore the establishment and maintenance of #kidney structure in mice…

New in the September issue of #G3journal: @yanjunzan and the team reanalyzed #transcriptome data from thousands of #maize to uncover the genetics underlying maize complex traits, and built a multi-tissue regulatory atlas for functional validation. buff.ly/bQMa6Gs

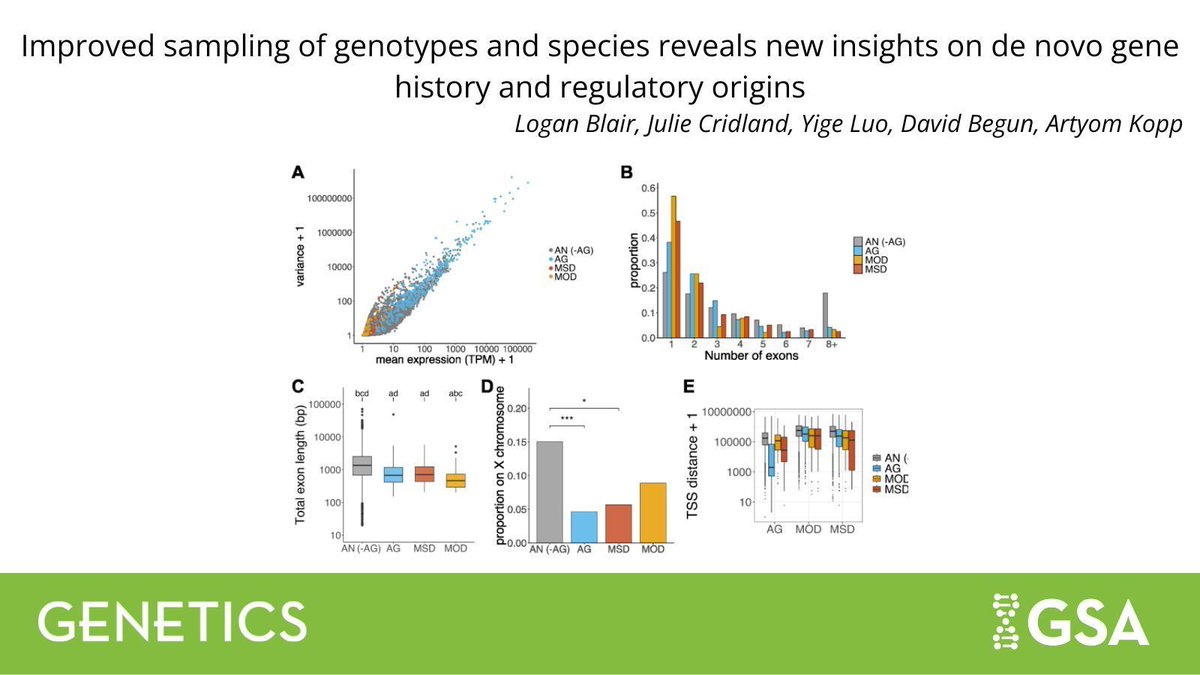
New research in #GENETICS provides new insights into the origin and early evolution of de novo genes by leveraging large-scale #transcriptome data from different #Drosophila genotypes. 🪰 Read more: buff.ly/Jwt8jjP
Join us @BioCAT_Poz & @UAM_IBMiB seminar by Evegenia Nitini's @evgenia_ntini from @IMBB_FORTH on the modelling of the nascent #RNA #transcriptome and resulting insights on processing of long non-coding RNAs and #enhancer transcription!

We found that a #biocontrol rhizobacterium responds to the #auxin indole-3-acetic acid by modifying its #transcriptome, increasing toxic compounds resistance, altering c-di-GMP levels and affecting #phage infection. @BlastMeetings @mSystemsJ @CSICdivulga journals.asm.org/doi/10.1128/ms…

Something went wrong.
Something went wrong.
United States Trends
- 1. New York 23.6K posts
- 2. New York 23.6K posts
- 3. Virginia 534K posts
- 4. Texas 226K posts
- 5. Prop 50 186K posts
- 6. #DWTS 41.2K posts
- 7. Clippers 9,683 posts
- 8. Cuomo 416K posts
- 9. TURN THE VOLUME UP 21.9K posts
- 10. Van Jones 2,512 posts
- 11. Ty Lue 1,006 posts
- 12. Harden 10K posts
- 13. Jay Jones 103K posts
- 14. Bulls 37.1K posts
- 15. #Election2025 16.5K posts
- 16. Sixers 13.1K posts
- 17. WOKE IS BACK 38.9K posts
- 18. Isaiah Joe N/A
- 19. #questpit 5,952 posts
- 20. Eugene Debs 3,191 posts