Cancer Data Science
@CancerDataSci
Fighting cancer with machine learning @ Broad Institute
You might like
While the focus in the clinical setting has been on finding DNA-based cancer biomarkers, in a study led by Josh Dempster we show that, at least in vitro, gene expression is generally a more powerful biomarker for cancer vulnerabilities biorxiv.org/content/10.110… @CancerDepMap
Congratulations to @corsellos on the Drug Repurposing paper published today @NatureCancer! An unexpectedly large number of non-oncology drugs selectively inhibit subsets of cancer cell lines: nature.com/articles/s4301…
We've just released the MIX-seq manuscript, data and code. Congrats James, Brent, Allie, Katie and the rest of the team for this cool work!
Excited to share our preprint describing MIX-seq [biorxiv.org/content/10.110…]! MIX-seq is an approach for multiplexed transcriptional profiling of chemical and genetic perturbations in large pools of co-treated cancer cell lines, with single-cell resolution. (1/5)
![CancerDepMap's tweet image. Excited to share our preprint describing MIX-seq [biorxiv.org/content/10.110…]! MIX-seq is an approach for multiplexed transcriptional profiling of chemical and genetic perturbations in large pools of co-treated cancer cell lines, with single-cell resolution. (1/5)](https://pbs.twimg.com/media/ELV_lnIXkAAS5xE.jpg)
REPURPOSING HUB DATA AND PREPRINT NOW OUT!! See how >4500 existing compounds performed when used across >550 cancer cell lines 💊 biorxiv.org/content/10.110…, data can be found on the #DepMap portal: depmap.org/portal/downloa…
19Q3 Cancer Dependency data now live! #CRISPR knockout screening data for 64 (😲!) new #cancer cell lines, plus accompanying gene expression, copy number and mutation profiles: depmap.org
Are the CRISPR screens published by Sanger today consistent with those done at Broad? We teamed up with @DepMapSanger to find out. The results are in: biorxiv.org/content/10.110… @CancerDepMap @biorxivpreprint #OpenScience
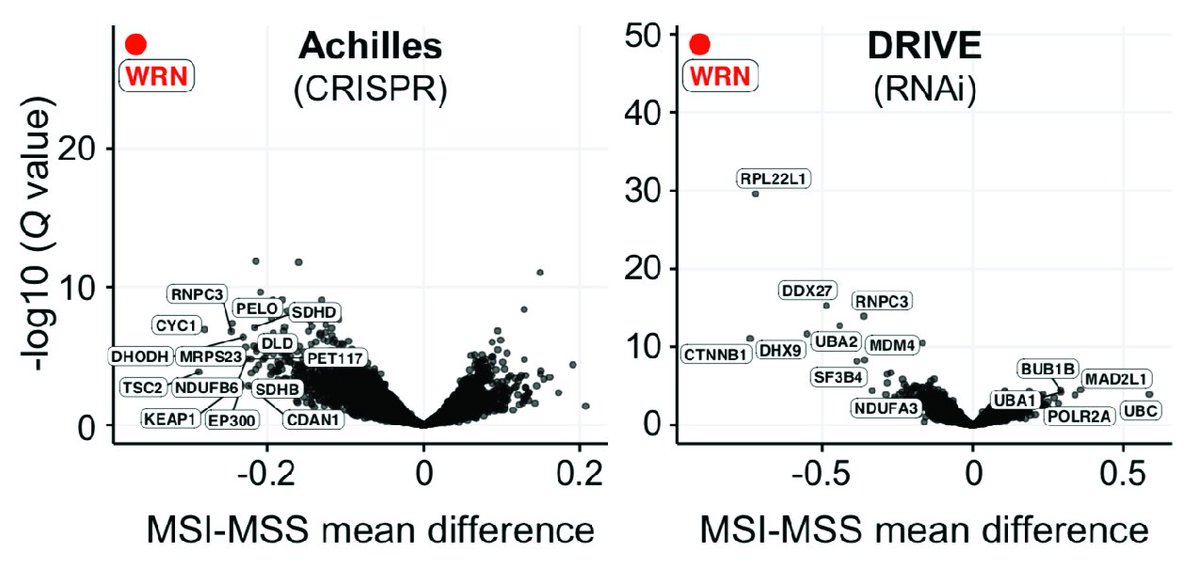
We’re excited to announce the release of a paper describing a new synthetic lethal target in MSI tumors as part of our GI Dependency Map! nature.com/articles/s4158… #Cancer #DepMap
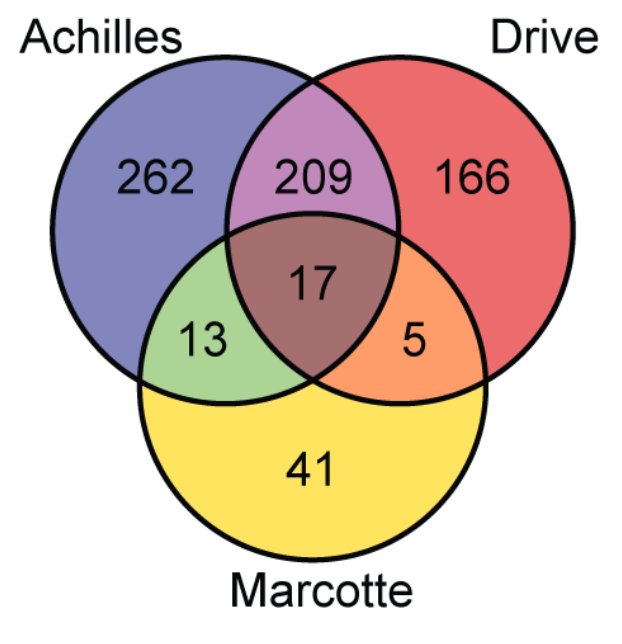
The largest dataset of cancer genetic dependencies to date includes RNAi data for 712 cell lines from Achilles, DRIVE and Marcotte et al., created using our new method DEMETER2, now out in Nat Comms. depmap.org/R2-D2 for data, code and paper @CancerDepMap @broadinstitute
A great example of how @CancerDepMap genetic screens can be used to study protein complex function. doi.org/10.1016/j.cels… Congrats to @joshbiology and @_robin_meyers_!
The largest dataset of cancer genetic dependencies: More than 970 RNAi screens in 713 cell lines from @broadinstitute Achilles, Novartis DRIVE and Marcotte et al., 2016 processed together by the new DEMETER2 method. biorxiv.org/content/early/… @CancerDepMap @biorxivpreprint
We have a new portal! depmap.org Integrated genome-scale #CRISPR LoF, sm mol sensitivity & #RNAi knockdown screen data on 500+ #cancer cell lines together w/ user-friendly analysis & #dataviz tools
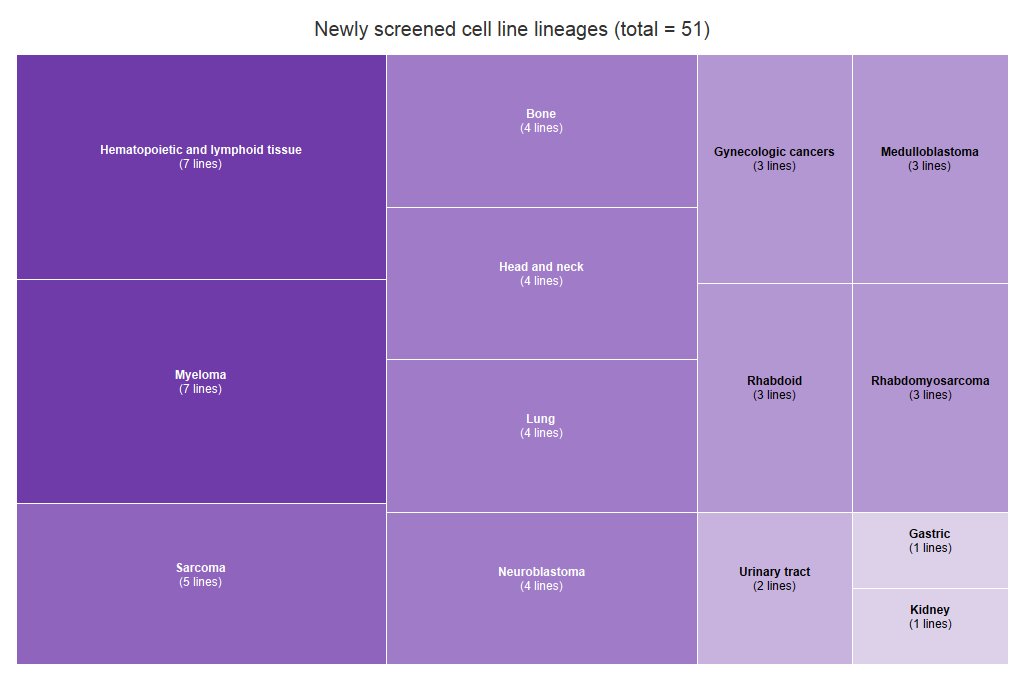
Now available from @CancerDepMap: #CRISPRscreen data from 391 #cancer lines, incl 51 new lines (representing 14 tumor types): broad.io/achilles Accompanying #RNAseq + #WES profiles: broad.io/CCLEdata. More to come on a quarterly basis!
342 CRISPR KO screens in cancer cell lines and our method to correct for CRISPR/CN effect, now live nature.com/ng/journal/vao… @broadinstitute
Our first effort to define a Cancer Dependency Map is now live at cell.com/cell/fulltext/… and depmap.org/rnai @broadinstitute
CERES corrects the copy-number effect of CRISPR-Cas9 essentiality screens in 342 cancer cell lines #bioRxiv biorxiv.org/content/early/…
United States Trends
- 1. South Carolina 28.1K posts
- 2. Texas A&M 27K posts
- 3. Shane Beamer 3,717 posts
- 4. Ryan Williams 1,241 posts
- 5. Michigan 44.1K posts
- 6. Sellers 13.8K posts
- 7. Northwestern 7,201 posts
- 8. Marcel Reed 4,715 posts
- 9. Ty Simpson 1,578 posts
- 10. Heisman 7,051 posts
- 11. #GoBlue 3,819 posts
- 12. Sherrone Moore 1,019 posts
- 13. Bryce Underwood 1,404 posts
- 14. Mateer 1,234 posts
- 15. Oklahoma 18.2K posts
- 16. College Station 2,933 posts
- 17. Nyck Harbor 3,352 posts
- 18. Andrew Marsh N/A
- 19. #GigEm 2,141 posts
- 20. Semaj Morgan N/A
You might like
Something went wrong.
Something went wrong.