#deconvolution search results
Evaluating performance and applications of sample-wise cell #deconvolution methods on human brain transcriptomic data biorxiv.org/content/10.110…

A labor of love: "Challenges and perspectives in computational deconvolution in genomics data". It discusses #deconvolution challenges across bulk measurement (gene expression and DNA #methylation) and #spatial #transcriptomics. #Bioinformatics arxiv.org/abs/2211.11808

Very excited to annouce the debut oral presentation from my new @UMich graduate student, Qianhui Huang, on Tues May 21st 2:10pm RNA II track, on evaluating #singlecell #deconvolution methods. She has the full spirit of new Garmire group. Thanks to @NIH @UMich and GLBio funding!

Join us on 6/19 at 10am to learn examples of the practical applications of #deconvolution from individual instruments to big clusters. #microscopy nvda.ws/2WDRDvx

Assessment of methods to deconvolve cellular composition from bulk gene expression. #DREAM, #Immunotherapy, #deconvolution nature.com/articles/s4146…

We are proud to announce that we have officially launched the Expansion Microscopy Studio (exm.studio), software designed to make life easier for researchers in biology labs across the world. highlights include GPU accelerated #deconvolution (the world's fastest!)

Our new preprint of "Deconvolution in Fluorescence Lifetime imaging microscopy (FLIM)" corrects the non-linear effects (blur and shift of ideal boundary) of convolution on lifetime in FLIM. (SPIE Photonics West conference) arxiv.org/pdf/2201.06136… #deconvolution #lifetime #FLIM

🚨Monday morning #microscopy PSA🚨 #Deconvolution is an extremely useful tool to improve your images but please, please, please do not deconvolve your PSF, that just doesn’t make any sense. david-hoffman.github.io/post/dont-deco…

Standard #deconvolution relies on a-priori assumptions that often produce artifacts. #TRUESHARP image boosting is better: It incorporates measured information from the image to reliably remove noise and background & enhance resolution without falsification. And now it’s…
Correction of Missing-Wedge Artifacts in Filamentous #Tomograms by Template-Based Constrained #Deconvolution pubs.acs.org/doi/10.1021/ac… #Current_issue #Bioinformatics

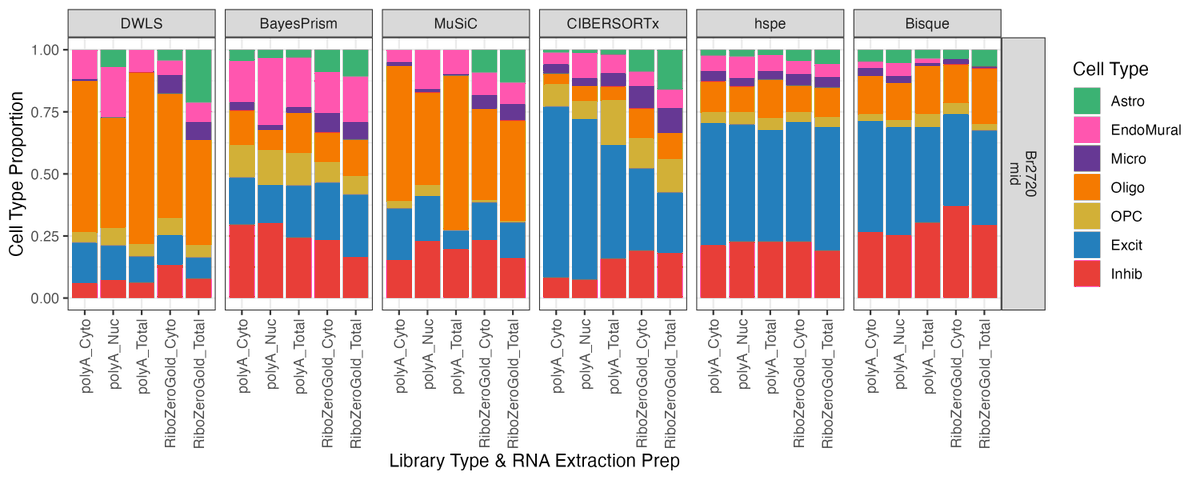
Hey #deconvolution fans! 👀 We’ve got an exciting new pre-print for you: a benchmark of 6 popular deconvolution methods on a multimodal human brain #DLPFC dataset 🧠🧬 @LieberInstitute @jhubiostat #scitwitter 📎 doi.org/10.1101/2024.0…
Interested in learning the basics of how you can apply real-time 3D #deconvolution to enhance fluorescence #microscopy? Join us for a webinar on 6/19. Register now: nvda.ws/2Icqzhu

And, no, not in #LCMS or #GCMS mass chromatograms one, can NOT assign "structures/ compounds/IDs" on "TICs"- fails the whole purpose of complex sample runs and data #deconvolution in #metabolomics. Its misleading representation. #massspec

The new freedom of image boosting: our #TRUESHARP online #deconvolution at app.truesharp.rocks! 1) open your image 2) adjust decon settings with just 4 sliders 3) enjoy intelligent image processing on your local device And yes, there is no need for registration and no…
When you don’t have a Thunder #microscope anymore but still need to improve your #confocal images. 🫠 #deconvolution

Click to try Nikon’s NIS-Elements software #deconvolution, for free, right now! Image stacks may be imported as ND2 or TIFF file types and up to 50 MB. bit.ly/2Gru5Vo


We added the automatic calculation of the SNR so that @svi_huygens adapts its #deconvolution for each #microscope #confocal #spinningdisk #Widefield images imported and saved in our #CochinImageDatabase Just a click away !!!
We will hold our first virtual Huygens workshop on April 14th & 15th. Participants get hands-on experience with #microscopy image restoration, visualization and analysis. There are a few spots left! To join, message info(at)svi.nl. #ImageAnalysis #Deconvolution

🔬 Choosing the right cellular deconvolution tool is key! With so many options, it’s crucial to match your tool to your needs! Read our blog to help you choose wisely! fiosgenomics.com/tool-selection… #Bioinformatics #RNAseq #Deconvolution #ComputationalBiology
⚠️ Is blur misleading your analysis? Deconvolution reverses that blur—sharpening images, revealing hidden structures, and helping you get results you can trust 👌 🔗 Full guide in comments #Deconvolution #Microscopy #ImageAnalysis #ImageProAI #LifeScience #MaterialsScience




1/3: 👏👏 We recommend an excellent tool, Deconer, a powerful reference-based deconvolution method for gene expression data. academic.oup.com/gpb/article/23… #deconvolution #gene_expression
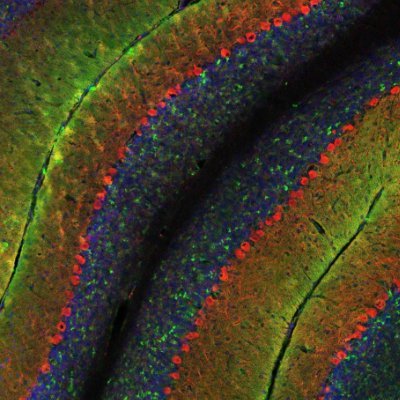
A wormhole in our universe? Animation of a curled up C. elegans nematode imaged with a spinning disk confocal, deconvolved and rendered using the Huygens Software. C/o Igor Bonacossa Pereira @HilliardLab, @QldBrainInst. #Neuroscience #microscopy #deconvolution #imageanalysis
Artery of an Atlantic salmon filled w/ nucleated red blood cells. Resseguir, 2022 #micro #Confocal #Deconvolution #Fluorescence

Artefact-free #deconvolution with abberior’s #TRUESHARP image boosting. Confocal image of the #Drosophila embryo CNS stained for tubulin (🟦STAR ORANGE) and DNA (🟩PicoGreen). TRUESHARP is also available as a free online tool: link.abberior.rocks/true #fluorescencefriday

Standard #deconvolution relies on a-priori assumptions that often produce artifacts. #TRUESHARP image boosting is better: It incorporates measured information from the image to reliably remove noise and background & enhance resolution without falsification. And now it’s…
The new freedom of image boosting: our #TRUESHARP online #deconvolution at app.truesharp.rocks! 1) open your image 2) adjust decon settings with just 4 sliders 3) enjoy intelligent image processing on your local device And yes, there is no need for registration and no…
🚀Going deeper, we demonstrate that #deconvolution is now a multi-modal data integration technique with most frameworks using #singlecell data, #spatial coordinates or tissue image. We delineate the diversity of approaches to integrate these data types.

We first present the overall workflow of cell-type #deconvolution for #spatialtranscriptomics from input to output. We highlight a first dichotomy between methods depending on wether they use #singlecell data and also pinpoint the novel use of tissue image as a new modality.

🔬 Choosing the right cellular deconvolution tool is key! With so many options, it’s crucial to match your tool to your needs! Read our blog to help you choose wisely! fiosgenomics.com/tool-selection… #Bioinformatics #RNAseq #Deconvolution #ComputationalBiology
Excited to announce that our #deconvolution benchmark has now been published in @GenomeBiology ! 📊 📄 doi.org/10.1186/s13059… Check out my blog post for a high level overview: lahuuki.github.io/posts/2025-04-…
Hey #deconvolution fans! 👀 We’ve got an exciting new pre-print for you: a benchmark of 6 popular deconvolution methods on a multimodal human brain #DLPFC dataset 🧠🧬 @LieberInstitute @jhubiostat #scitwitter 📎 doi.org/10.1101/2024.0…

See the difference Huygens deconvolution can make! Reveal the hidden details in your images with just a few clicks, and efficiently process all your images in batches. Get in touch to try it out for free on your own data at LCIF! #Microscopy #Deconvolution #Huygens
Matrix STED image reveals detail of Nuclear pore complex protein fluorescence using @abberior STAR-RED after Huygens unique SuperXY #deconvolution. Thanks to Marie Olšinová & Aleš Benda, Imaging Methods Core Facility – BIOCEV @CharlesUniPRG for providing the raw data
The new freedom of image boosting: 1) open your image 2) adjust decon settings with just 4 sliders 3) enjoy intelligent image processing on your local device That’s #TRUESHARP online #deconvolution! And yes, there is no need for registration and no fee 😊 app.truesharp.rocks
ICYMI: A new tool called MUSTANG is making waves in #spatial genomics research by enabling researchers to go beyond #scRNAseq analysis and explore multi-sample insights, impossible just months ago. Read more on our blog: bridgeinformatics.com/cross-sample-a… #Deconvolution #geneexpression

Do you know what's SMART? SMART=Spatial transcriptomics deconvolution using MARker-gene-assisted Topic model. It is a novel #deconvolution algorithm to take full advantage of #spatial #omics data. A beautiful work by our PhD student, Yolanda Yang. rdcu.be/d2fMb

A labor of love: "Challenges and perspectives in computational deconvolution in genomics data". It discusses #deconvolution challenges across bulk measurement (gene expression and DNA #methylation) and #spatial #transcriptomics. #Bioinformatics arxiv.org/abs/2211.11808

Evaluating performance and applications of sample-wise cell #deconvolution methods on human brain transcriptomic data biorxiv.org/content/10.110…

Excited to have submitted a poster abstract on deconvolution in brain tissue to @ACNPorg ! 🧠 😎🧪#deconvolution

We are proud to announce that we have officially launched the Expansion Microscopy Studio (exm.studio), software designed to make life easier for researchers in biology labs across the world. highlights include GPU accelerated #deconvolution (the world's fastest!)

Rolando Berlinguer Palmini & Matthew Zorkau (@UniofNewcastle) used Huygens #deconvolution and Huygens Surface Renderer to show newly synthesised mitochondrial protein. Below an animated version of their #nanoscopy-resolution image, 3rd prize in the Huygens Image Contest
Click to try Nikon’s NIS-Elements software #deconvolution, for free, right now! Image stacks may be imported as ND2 or TIFF file types and up to 50 MB. bit.ly/2Gru5Vo

📢 Our review on next-generation #deconvolution of the tumor microenvironment has been published on @IRCellMolecBio! 🎉 shorturl.at/hlnX7 Kudos to @lorenzo_merotto @mzopog and @ConstantinZackl! @DiSC_uibk @uniinnsbruck

We will hold our first virtual Huygens workshop on April 14th & 15th. Participants get hands-on experience with #microscopy image restoration, visualization and analysis. There are a few spots left! To join, message info(at)svi.nl. #ImageAnalysis #Deconvolution

How to get rid of background and noise while enhancing resolution? With #TRUESHARP image boosting! TRUESHARP is intelligent #deconvolution you can trust: It doesn’t work with assumptions but uses information from the sample to remove noise and background. Learn more:…

Standard #deconvolution relies on a-priori assumptions about sample structure that often produce artifacts. #TRUESHARP image boosting is better. It incorporates measured information from the image to reliably remove noise and background & enhance resolution without falsification.…


⚠️ Is blur misleading your analysis? Deconvolution reverses that blur—sharpening images, revealing hidden structures, and helping you get results you can trust 👌 🔗 Full guide in comments #Deconvolution #Microscopy #ImageAnalysis #ImageProAI #LifeScience #MaterialsScience




Very excited to annouce the debut oral presentation from my new @UMich graduate student, Qianhui Huang, on Tues May 21st 2:10pm RNA II track, on evaluating #singlecell #deconvolution methods. She has the full spirit of new Garmire group. Thanks to @NIH @UMich and GLBio funding!

Live talk during Expo#3 Today at 9:45AM (PST) @AIM2021 @BerkeleyMIC. Learn to get amazing results from a Widefield (@DeltaVisionOMX) or ANY other microscope type with Huygens #deconvolution, visualization & #imageanalysis. c/o: Dr. Alexia Ferrand, ICF, @biozentrum, @UniBasel_en
Celebrating your #ScienceArt! Join the Huygens #Microscopy Image Contest and win a laptop! More info: svi.nl/ImageContest #deconvolution #imageanalysis #cellbiology #superresolution #bioart @AmyEngevik @MicroMindy

1000 Twitter followers! Keeping in touch with our clients and the general research field is an important part of our company's mission. Connecting with more of you through social media is therefore cause for celebration! #Deconvolution #ImageAnalysis #Microscopy #Huygens

One-step #deconvolution for #multi-#angle #TIRF #microscopy with enhanced #resolution Junchao Fan, Xiaoshuai Huang, Liuju Li, Liangyi Chen, and Shan Tan doi.org/10.1364/BOE.10…

Image #deconvolution for confocal laser scanning #microscopy using constrained total variation with a gradient field Tao He, Yasheng Sun, Jin Qi, Jie Hu, and Haiqing Huang doi.org/10.1364/AO.58.…

#OES_highlight Deblurring, artifact-free optical coherence tomography with deconvolution-random phase modulation doi.org/10.29026/oes.2… by Prof. #LinboLiu from @NTUsg #deconvolution #deblurring #OCT #tomography #PhaseModulation


Join us on 6/19 at 10am to learn examples of the practical applications of #deconvolution from individual instruments to big clusters. #microscopy nvda.ws/2WDRDvx

Something went wrong.
Something went wrong.
United States Trends
- 1. Raindotgg 1,124 posts
- 2. Louisville 14K posts
- 3. Ortiz 15.8K posts
- 4. #GoAvsGo 1,427 posts
- 5. Nuss 5,488 posts
- 6. Batum N/A
- 7. Emmett Johnson 2,031 posts
- 8. Miller Moss 1,167 posts
- 9. UCLA 7,491 posts
- 10. #Huskers 1,038 posts
- 11. #FlyTogether 1,587 posts
- 12. Bama 13.5K posts
- 13. Oilers 4,323 posts
- 14. #Mashle N/A
- 15. Brohm 1,133 posts
- 16. #TheFutureIsTeal 1,042 posts
- 17. Nikki Glaser N/A
- 18. Lateef 2,161 posts
- 19. The ACC 19.7K posts
- 20. Ty Simpson 3,533 posts