#spatialtranscriptomics 검색 결과
#SpatialTranscriptomics Visium @10xGenomics Tuberous sclerosis complex-associated Renal Angiomyolipoma (n=1) vs sporadic AML (n=1) vs Renal Cell Carcinoma (n=1) Melanocytic traits in AML MITF MLANA PMEL @RyutaWatanab999 @SciReports 2025 nature.com/articles/s4159…

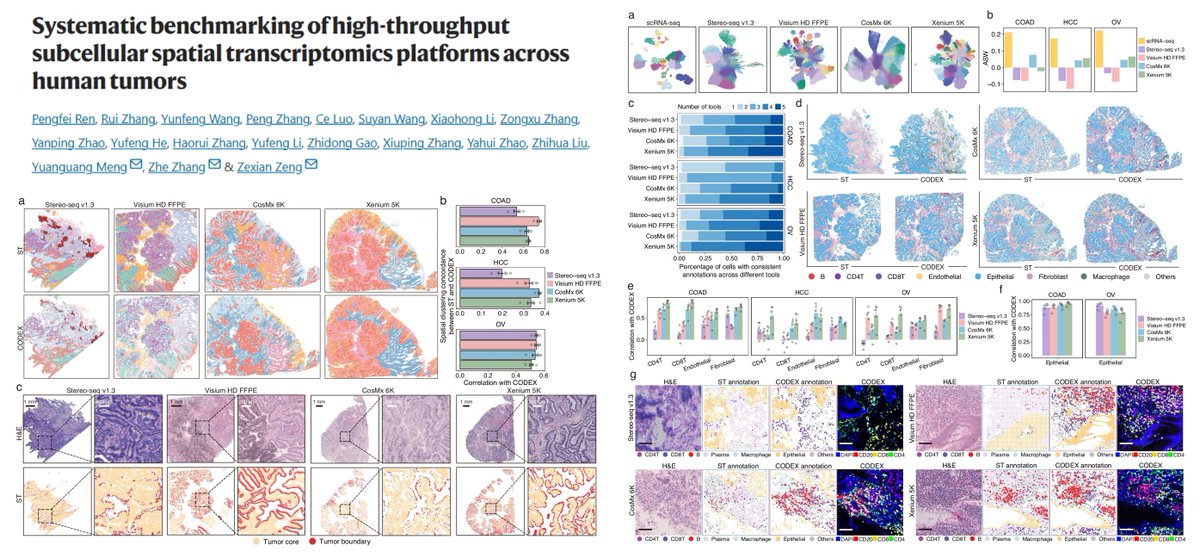
Systematic comparisons among 4 subcellular-resolution #SpatialTranscriptomics methods Stereo-seq v1.3 Visium HD FFPE CosMx 6k Xenium 5k Human Tumors Ground truth CODEX, +scRNAseq Gene detection sensitivity (CosMx performance...🙁) (Spatial) false positives Transcript-protein…
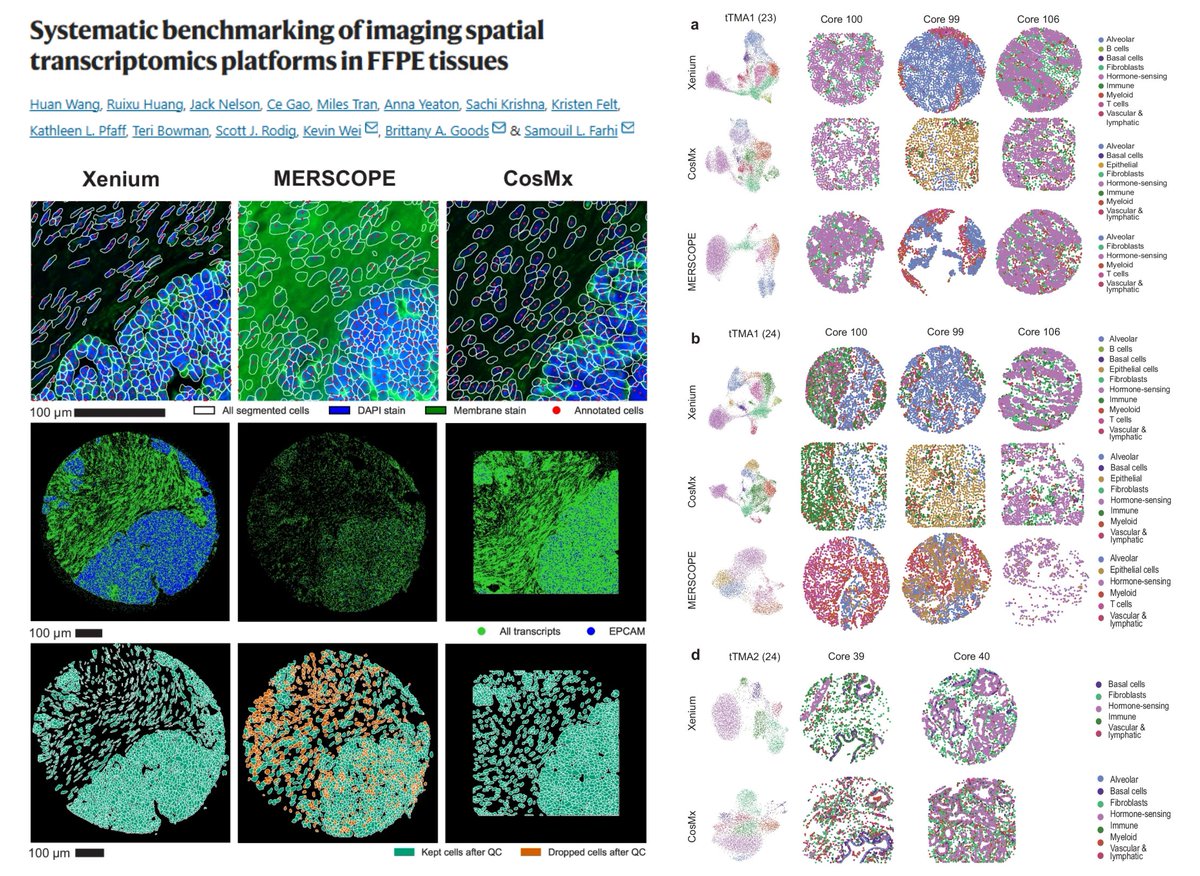
Comparative analysis of commercial Imaging-based #SpatialTranscriptomics methods Xenium CosMx 1k MERSCOPE Tissue Microarray 17 tumors + 16 normal tissues Transcript count Corr. with reference bulk TCGA GTEx RNAseq or scRNAseq Segmentation (Precision Recall F1 score) Accuracy:…
Congrats to Dr. Savannah LaBuda on being one of the winners of the Young Investigator Poster Award at #AMPath2025! 🎉👏 #SpatialTranscriptomics #GYNpath #MolPath @AMPath @pvhernandezmd @whosainastro

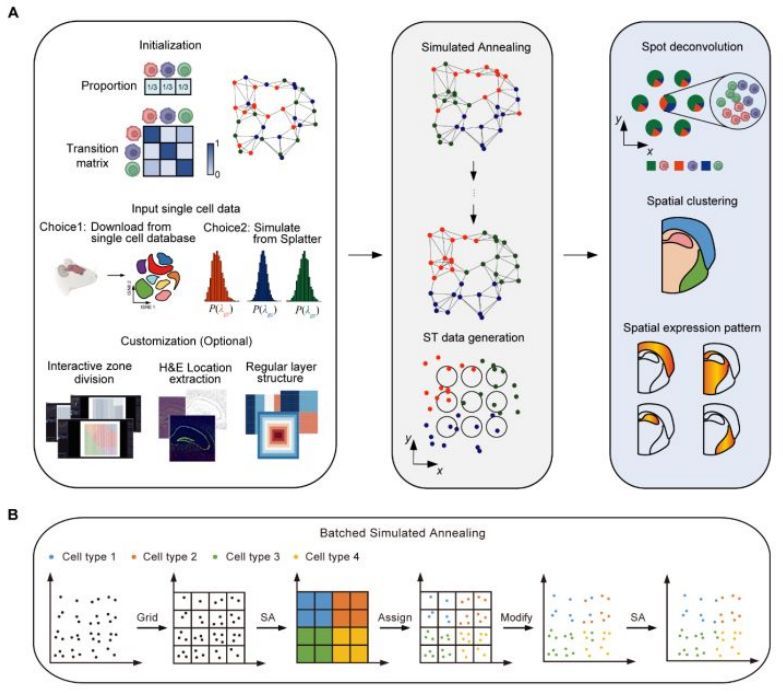
Spider: a flexible and unified framework for simulating spatial transcriptomics data. #SpatialTranscriptomics #DataSimulation #Genomics #Bioinformatics 🧬 🖥️ academic.oup.com/bioinformatics…
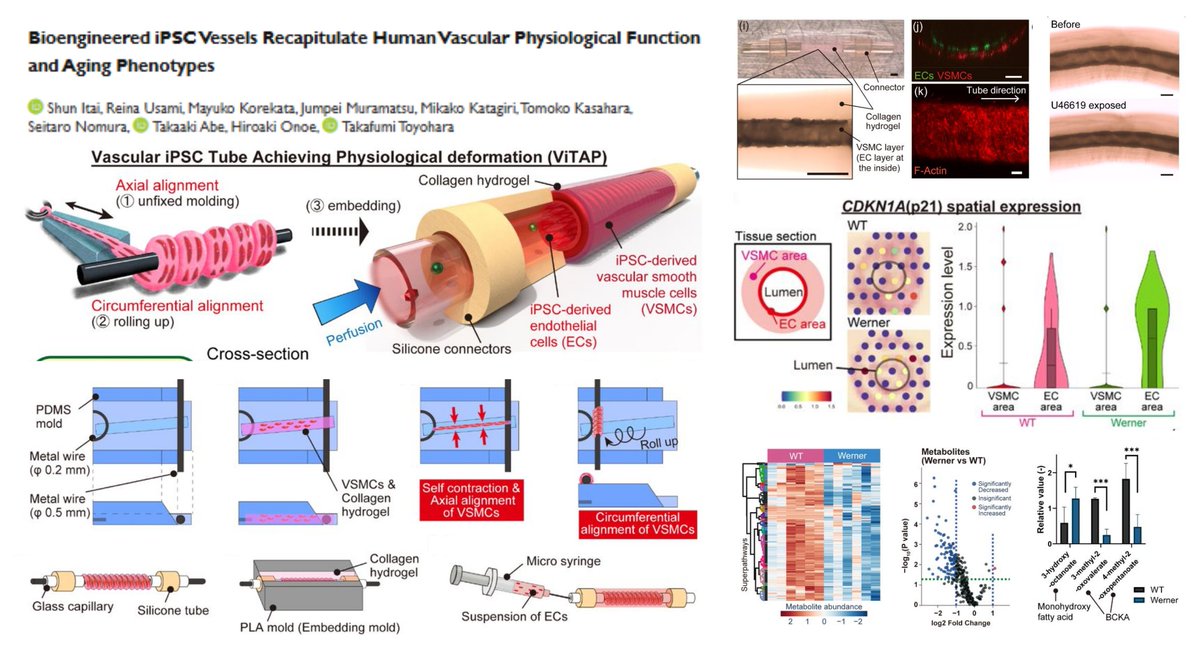
#WernerSyndrome hiPSC-derived engineered vessel #SpatialTranscriptomics #Visium @10xGenomics + #Metabolomics▶️ #BranchedChainKetoAcid⬇️ #MonohydroxyFattyAcid⬆️ in Werner vessels with traits of #PrematureAging Aligned #SmoothMuscleCell in Collagen hydrogel➡️ Roll-up around φ…
"We all love the pretty pictures we get from #SpatialTranscriptomics but the real value lies in the ability to precisely determine the cellular composition & cell-cell relationships - not possible with bulk RNA or even scRNA approaches" 💯% well said @bayraktar_lab! 👏 #FoG2025




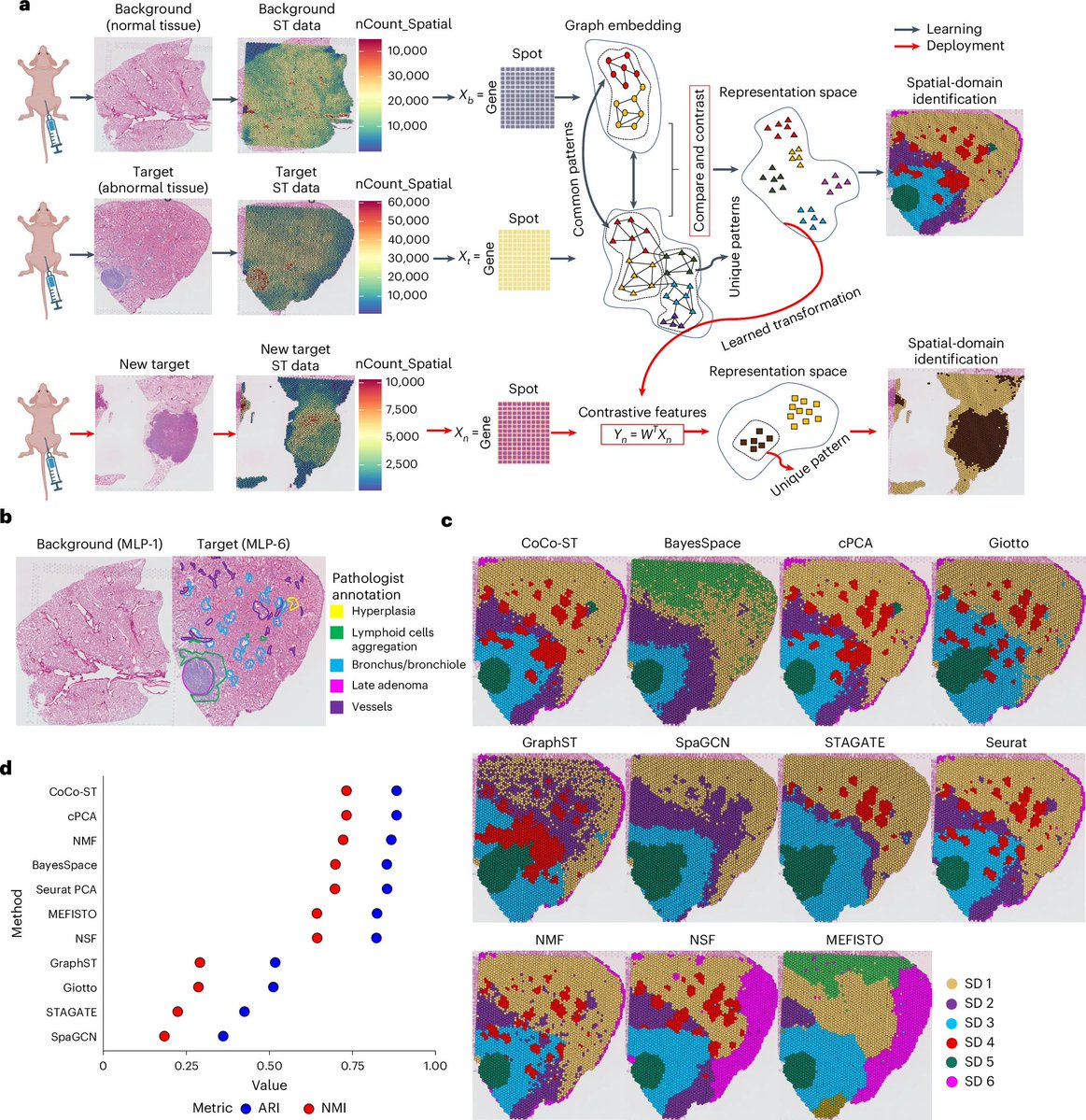
CoCo-ST detects global and local biological structures in spatial transcriptomics datasets. #SpatialTranscriptomics #VarianceIdentification @NatureCellBio nature.com/articles/s4155…
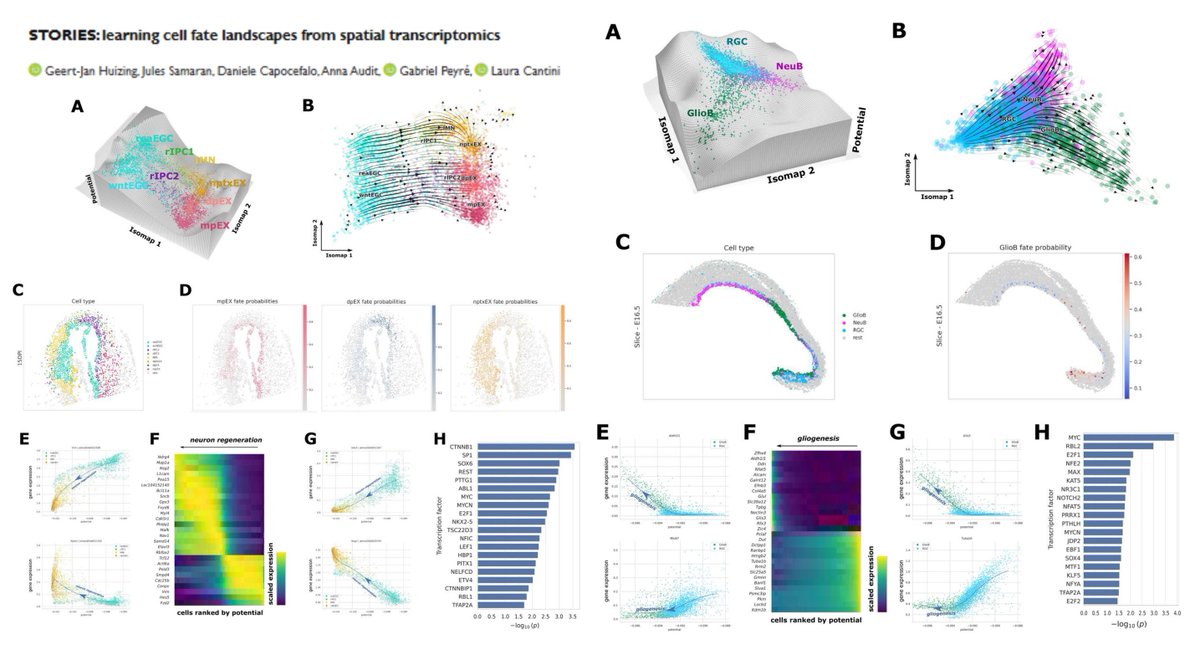
STORIES SpatioTemporal Omics eneRgIES Learning Waddington epigenetic (+regulon) landscape from multi-time-point Stereo-seq #SpatialTranscriptomics Potential-based Fused Gromov-Wasserstein #OptimalTransport Linear▶️gene expression error Quadratic▶️spatial consistency @gjhuizing…
GOD IS GOOD! Today, I passed my #PhD Dissertation Defense!!! 🎉 Thank you to everyone who believed in me. Thank you to all my mentors, friends, and family. 🥰 I would not be where I am today without all of you. 🙏❤️ @SBS_UNTHSC #SpatialTranscriptomics #DoubleDocs @A_P_S_A


#SpatialOmics #ImagingMassSpec #SpatialTranscriptomics MISTy Multiview Intercellular SpaTial modeling framework bioconductor.org/packages/relea… View/spatial scale-specific feature interactions Intraview - same location (intra-to-multicellulat) Juxtaview - immediate neighborhood…

Excellent talk from @Sandy_Figiel in @Uroweb #EAUlab session at #EAU25 presenting our cool data (I'm bias!) on #SpatialTranscriptomics & #ClonalAnalysis to detect unique features of metastasing clones in #ProstateCancer. @OxPCaBiol #SPACE_Study #3DLightsheet




Comparing single-cell #SpatialTranscriptomics methods on FFPE human tumor #TissueMicroArray CosMx 1k vs MERFISH 500 vs Xenium-UM/-MM 339 93 common genes lung adenocarcinoma pleural mesothelioma Corr. Bulk RNAseq, GeoMx "better F1-scores with Xenium-MM (median, > 75%) than…

Are you curious to know how using #SpatialTranscriptomics we have discovered a new organ in the 21th century? For this, an oral-gut neural physiological circuit and more you can visit my poster P2.21.23 at #ECI2024 poster session :) @y_efis @EFIS_Immunology

#SpatialTranscriptomics Visium #CerebralMalaria 🐭+P.berghei ANKA -/+Artesunate Malaria infection▶️ ⏫🧠#EndothelialCell, immune cells SPATA spatial trajectory inference of EC-rich spots in infected🧠▶️ ⏫EC antigen processing/presentation + leukocyte adhesion ⏬BBB Partially…

Ruochen Dong, @linheng_li lab, gave an outstanding presentation on using #SpatialTranscriptomics to interrogate HSC-niche interaction in the mouse fetal liver at #ISSCR2023. He also received merit and travel awards. Congratulations! @ISSCR

⚡️Exciting Breakthrough in Progressive Multiple Sclerosis⚡️ Much humbled to contribute to this exciting project from @Pluchinolab . Analysing 200+ sections from 50+ patients, showcases the power of #spatialtranscriptomics in rare cell localization. 🧠 @IBT_CAS @LukasValihrach
Proud to see our work out in @NeuroCellPress ! cell.com/neuron/fulltex… With Bongsoo Park, Dimitris Tsitsipatis @DanielZucha @LukasValihrach @irina_mohorianu @Pluchinolab @BeermanHSCaging !
Something went wrong.
Something went wrong.
United States Trends
- 1. Bama 48.1K posts
- 2. Georgia 61.4K posts
- 3. Ty Simpson 7,277 posts
- 4. Miami 226K posts
- 5. #NXTDeadline 13K posts
- 6. #UFC323 24.5K posts
- 7. #SECChampionship 5,247 posts
- 8. #GoDawgs 14.3K posts
- 9. Ryan Williams 2,765 posts
- 10. DeBoer 4,368 posts
- 11. Frank Martin 1,880 posts
- 12. Kirby 15.5K posts
- 13. Texas Tech 32.3K posts
- 14. Jalin Turner 1,429 posts
- 15. Grubb 2,375 posts
- 16. #RollTide 3,064 posts
- 17. Oba Femi 7,126 posts
- 18. Messi 318K posts
- 19. Fowler 1,945 posts
- 20. Vettori 1,109 posts